Lax 33014 elife-33014-fig4-figsupp3-v1
Description:
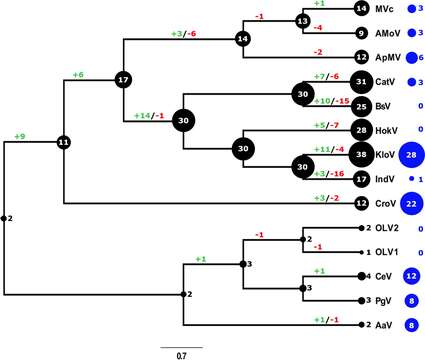
Description: English: Evolutionary history of translational machinery found in giant viruses inferred by COUNT and abundance of ankyrin repeat-domain genes. The size of the black circles mapped on a cladogram of members of the Mimiviridae represents the number of gene families involved in translation at each node or tip. Blue circles indicate the number of ankyrin repeat-domain encoding genes found in each genome. Gene gain and loss events are depicted along the branches. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus. Date: 27 March 2018. Source: https://elifesciences.org/articles/33014/figures at https://elifesciences.org/articles/33014 The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. In: eLife 2018;7:e33014 doi:10.7554/eLife.33014 . Author: Christoph M. Deeg, Cheryl-Emiliane T. Chow, Curtis A. Suttle. Other versions: .
Included On The Following Pages:
This image is not featured in any collections.
Source Information
- license
- cc-by-sa-3.0
- copyright
- Christoph M. Deeg, Cheryl-Emiliane T. Chow, Curtis A. Suttle
- creator
- Christoph M. Deeg, Cheryl-Emiliane T. Chow, Curtis A. Suttle
- source
- https://elifesciences.org/articles/33014/figures at https://elifesciences.org/articles/33014 The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. In: eLife 2018;7:e33014 doi:10.7554/eLife.33014
- original
- original media file
- visit source
- partner site
- Wikimedia Commons
- ID