Fmicb-09-01941-g002
Description:
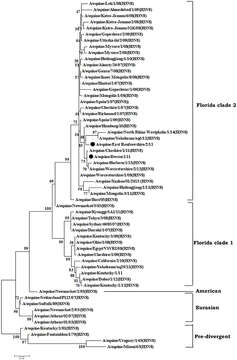
Description: English: Phylogenetic analysis of hemagglutinin (HA) genes nucleotide sequences from 57 Equine Influenza Viruses (EIVs). The maximum likelihood tree was constructed using stringent T92 + G algorithm which was identified using the find best DNA/protein model tool available in MEGA 6. The reliability of the trees was assessed by bootstrap with 1,000 replications with cut off at 50 are shown in the tree. The phylogram depicts five major clusters of global EIVs. Phylogenetic group's viz., Florida sub-lineage clade 1, Florida sub-lineage clade 2, American, Eurasian and Pre-divergent, are mentioned by bars on the right. The major mutations (I179V and A144V) observed in the Clade 2 viruses of Florida sublineage in recent isolates have been denoted by solid dots. Date: 6 September 2018. Source: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01941/full. Author: Raj K. Singh, Kuldeep Dhama, Kumaragurubaran Karthik, Rekha Khandia, Ashok Munjal, Sandip K. Khurana, Sandip Chakraborty, Yashpal S. Malik, Nitin Virmani, Rajendra Singh, Bhupendra N. Tripathi, Muhammad Munir, and Johannes H. van der Kolk.
Included On The Following Pages:
This image is not featured in any collections.
Source Information
- license
- cc-by-3.0
- copyright
- Raj K. Singh, Kuldeep Dhama, Kumaragurubaran Karthik, Rekha Khandia, Ashok Munjal, Sandip K. Khurana, Sandip Chakraborty, Yashpal S. Malik, Nitin Virmani, Rajendra Singh, Bhupendra N. Tripathi, Muhammad Munir, and Johannes H. van der Kolk
- creator
- Raj K. Singh, Kuldeep Dhama, Kumaragurubaran Karthik, Rekha Khandia, Ashok Munjal, Sandip K. Khurana, Sandip Chakraborty, Yashpal S. Malik, Nitin Virmani, Rajendra Singh, Bhupendra N. Tripathi, Muhammad Munir, and Johannes H. van der Kolk
- source
- https://www.frontiersin.org/articles/10.3389/fmicb.2018.01941/full
- original
- original media file
- visit source
- partner site
- Wikimedia Commons
- ID