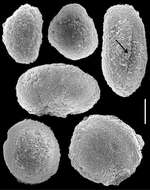
-
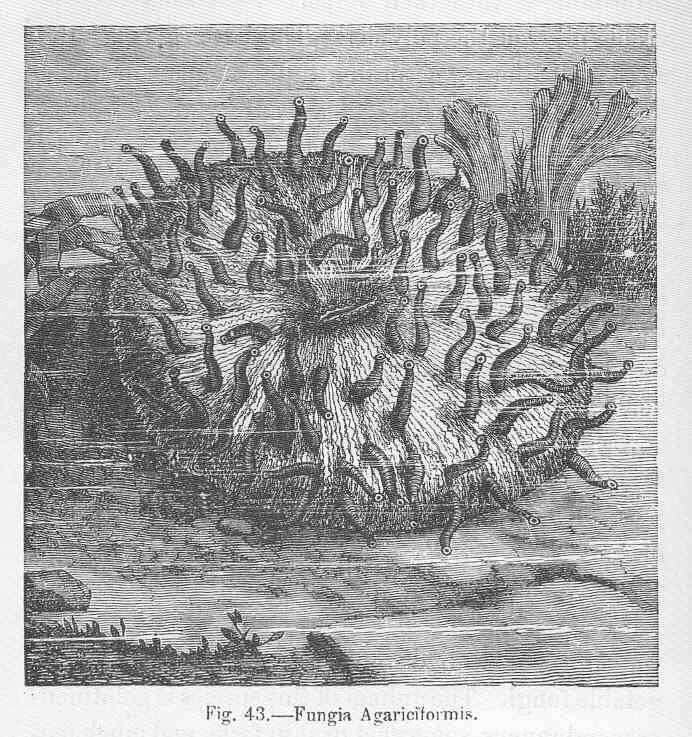
Fungia agariciformis.
-
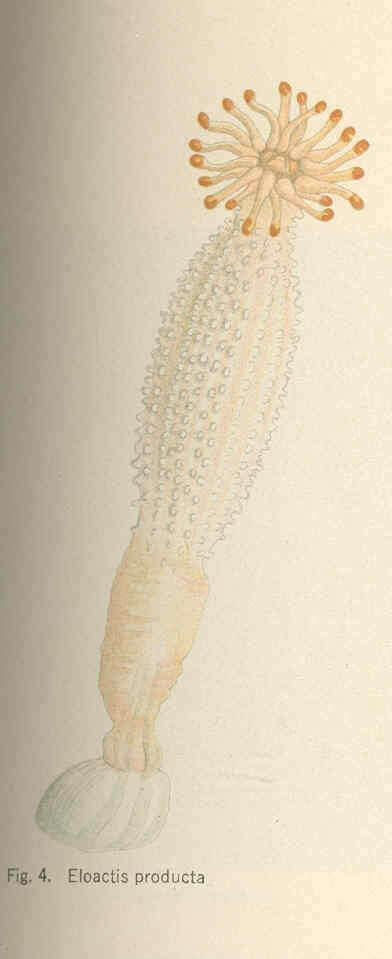
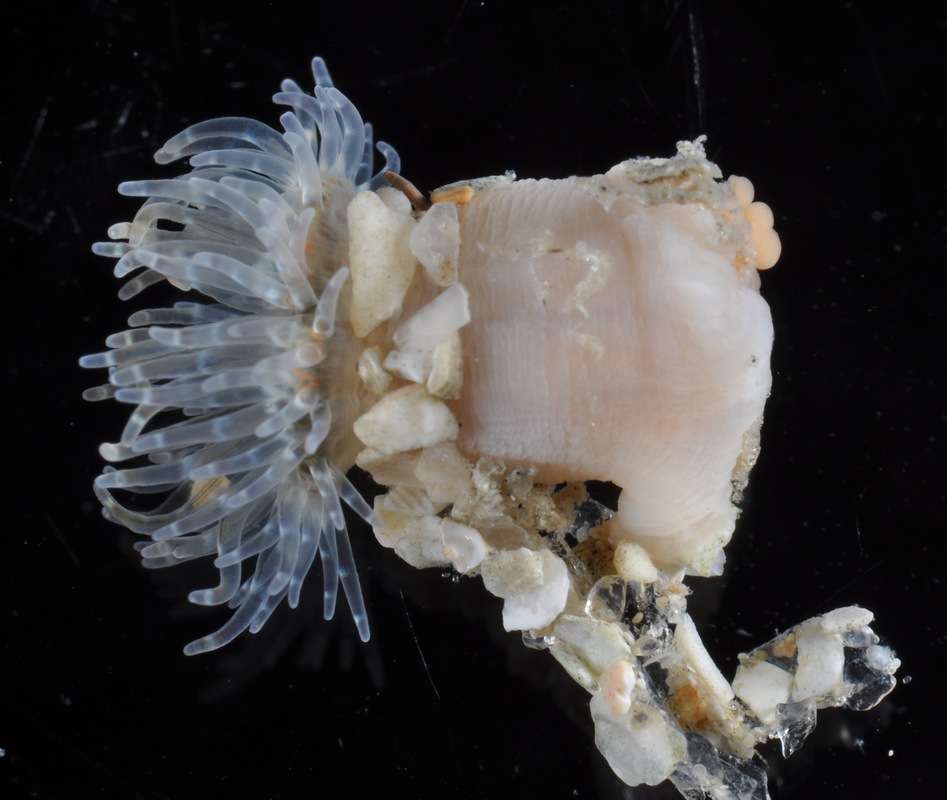
Eloactis producta.
-
-
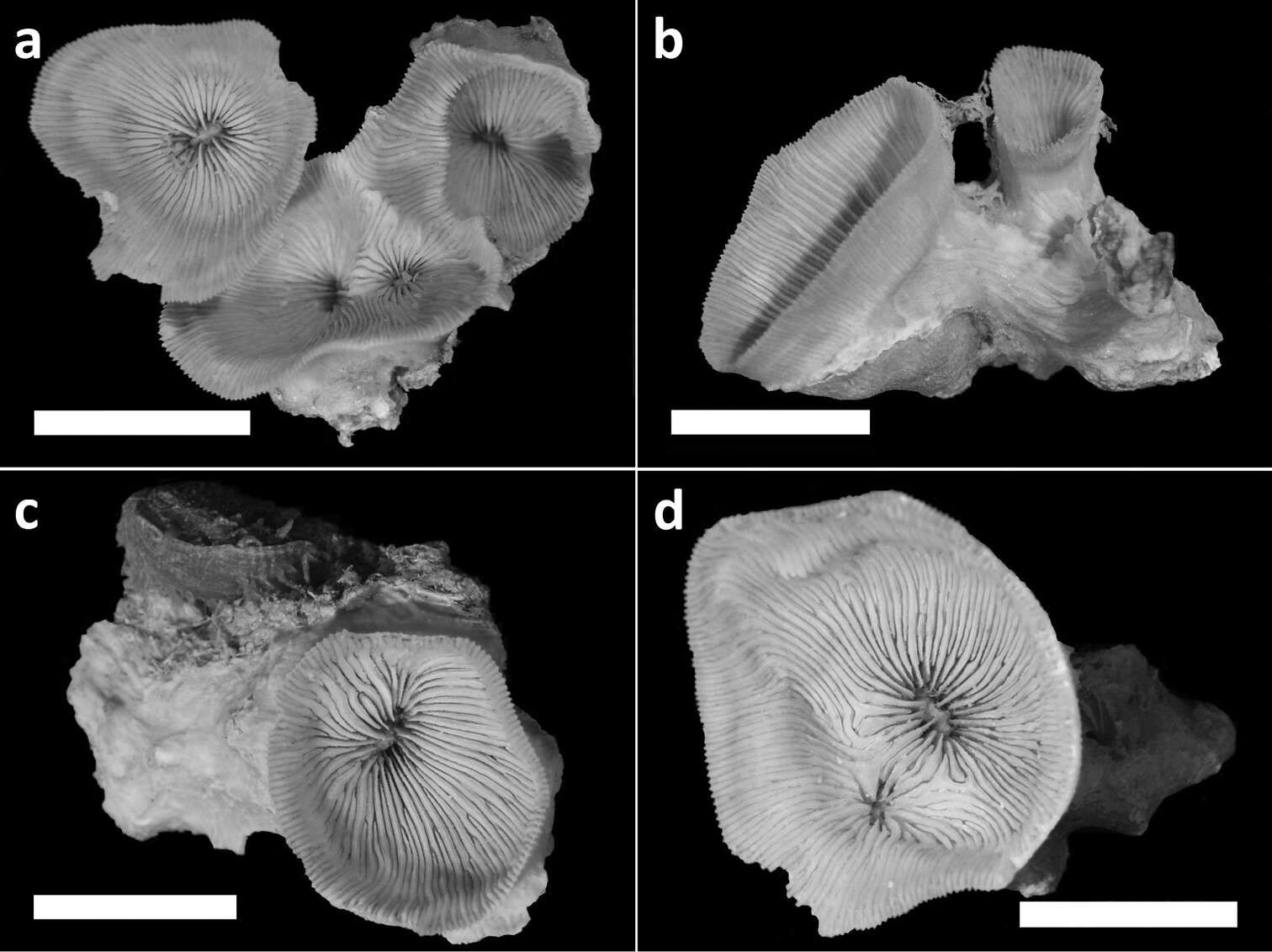
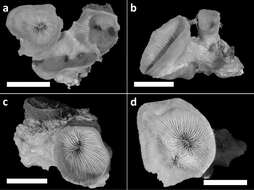
Figure 3.Holotype (RMNH Coel. 40138) and three paratypes (RMNH Coel. 40139) of Leptoseris troglodyta sp. n.from Palau. Scale bars: 1 cm. a Holotype consists of four calices: one (most left) has fused mid-height its calyx with two totally fused calices (centre), while another (right) has fused only with its corallum margin to those at the centre and the rest of its calyx has remained separate b Paratype: two separate calyces c Paratype: single calyx d Paratype: two fused calices. Scale bars: 1 cm.
-
Leen P. van Ofwegen, Yehuda Benayahu, Catherine S. McFadden
Zookeys
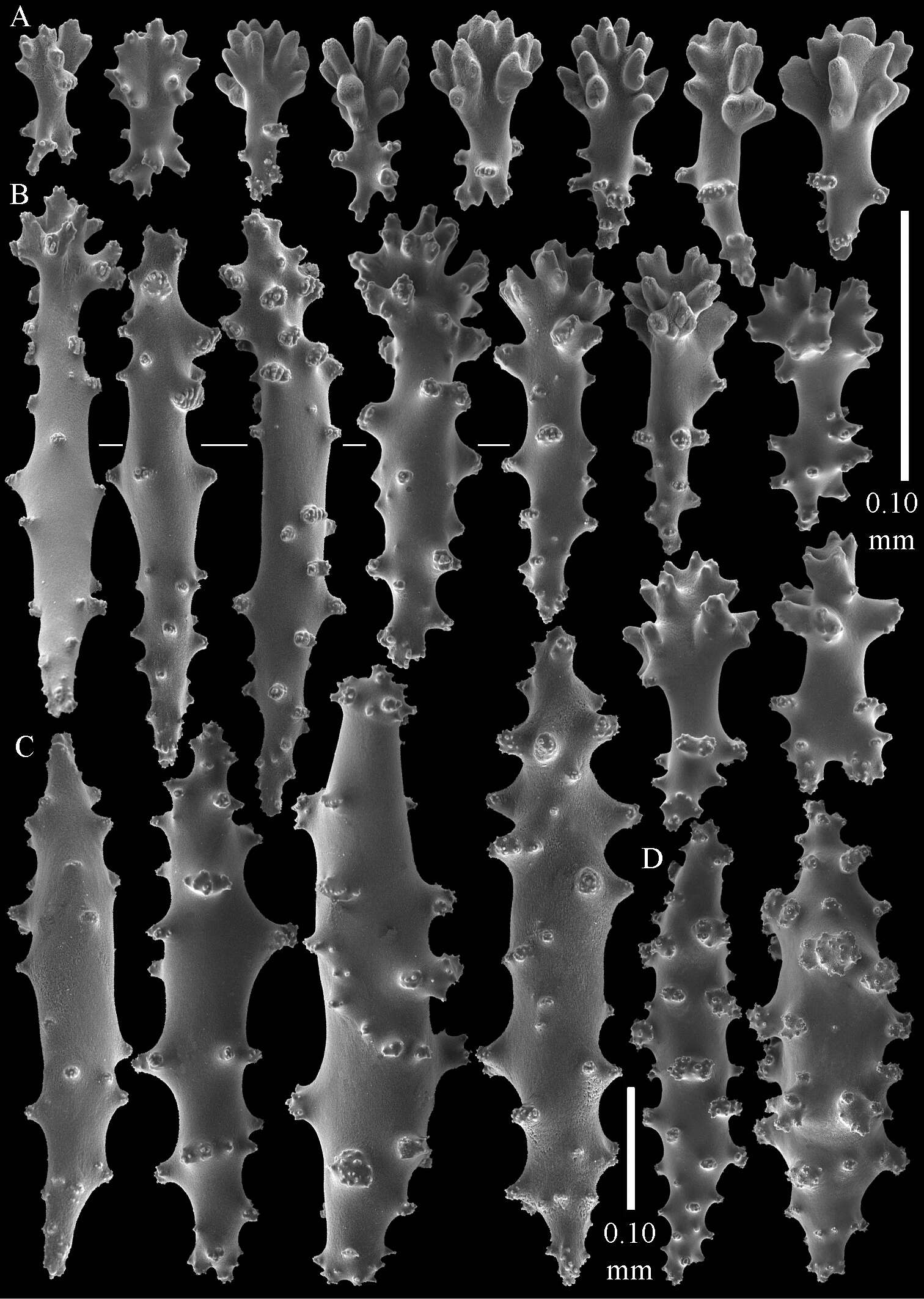
Figure 3.Sinularia australiensis sp. n., holotype NTM C14519. Sclerites of the surface layer of the base of the colony A leptoclados-type clubs B wart clubs C–D spindles. Scale of 0.10 mm at D only applies to D.
-
Ricardo González-Muñoz, Nuno Simões, José Luis Tello-Musi, Estefanía Rodríguez
Zookeys
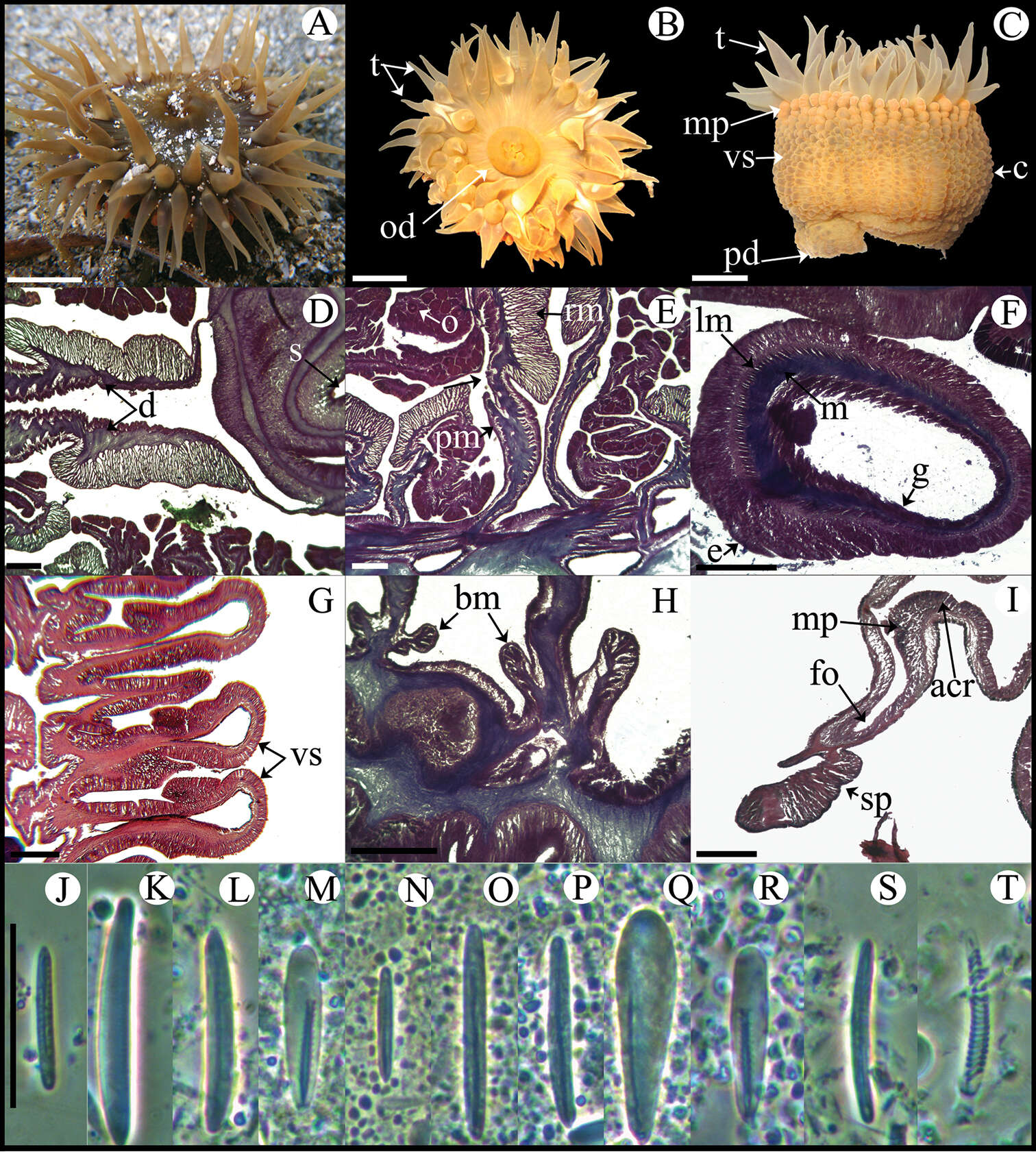
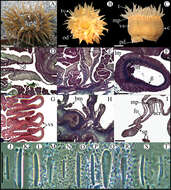
Figure 5.Bunodosoma cavernatum. A Live specimen in natural habitat B Oral view C Lateral view D Detail of directives; notice siphonoglyph E Cross section through proximal column showing oocytes F Cross section through tentacle G Longitudinal section through column showing vesicles H Longitudinal section though base showing basilar muscles I Longitudinal section through margin showing acrorhagi and marginal sphincter muscle J–T Cnidae.– acrorhagi: J basitrich K holotrich; actinopharynx: L basitrich M microbasic p-mastigophore; column: N small basitrich O basitrich; filament: P basitrich Q microbasic b-mastigophore R microbasic p-mastigophore; tentacle S basitrich T spirocyst. Abbreviations.– acr: acrorhagi, bm: basilar muscle, c: column, d: directives, e: epidermis, fo: fosse, g: gastrodermis, lm: longitudinal muscles, m: mesoglea, mp: marginal projection, o: oocyst, od: oral disc, pd: pedal disc, pm: parietobasilar muscle, rm: retractor muscle, s: siphonoglyph, sp: sphincter, t: tentacle, vs: vesicles. Scale bars: A–C: 10 mm; D–I: 200 μm; J–T: 25 μm.
-
Catherine S. McFadden, Leen P. van Ofwegen
Zookeys
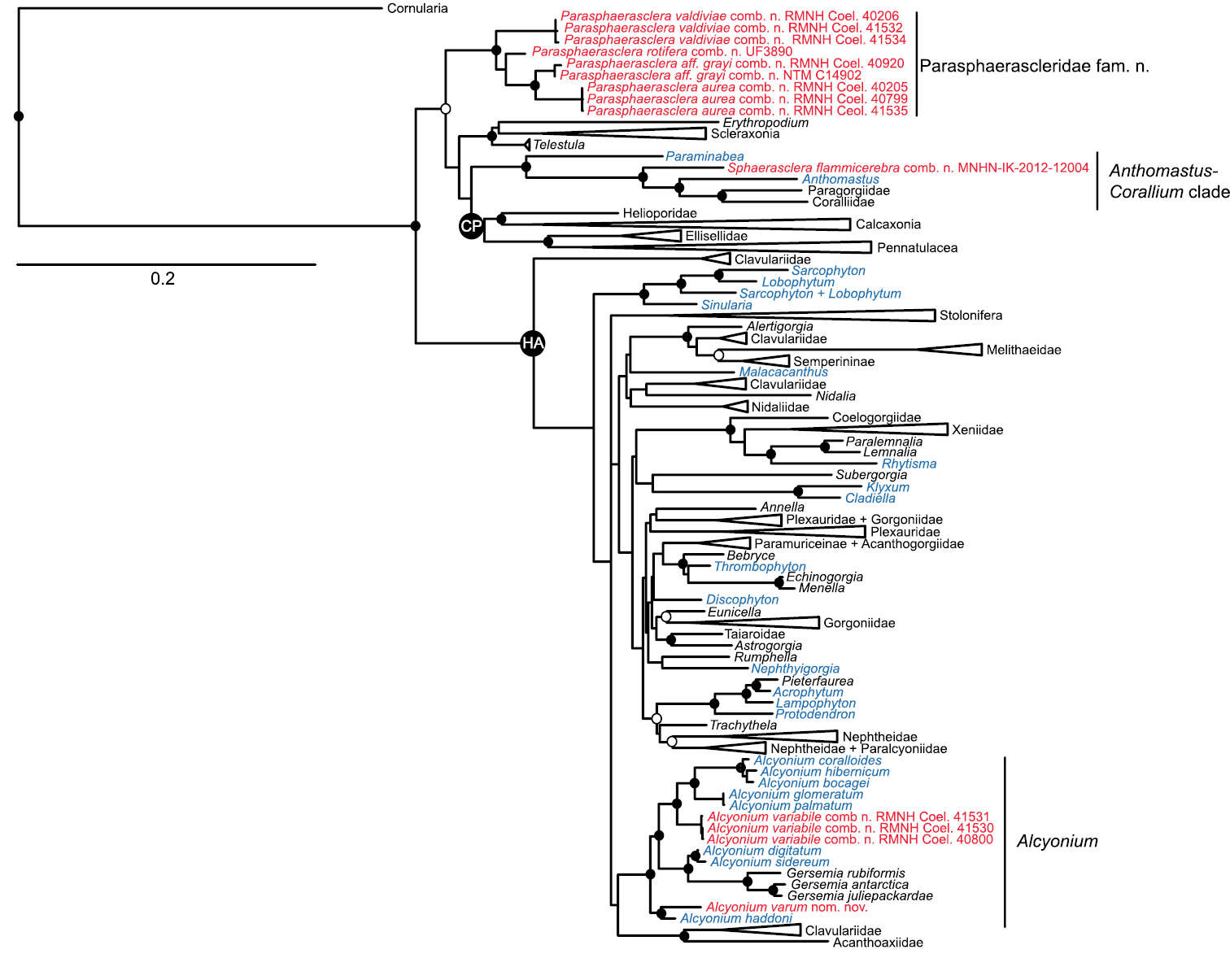
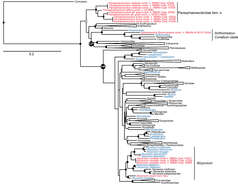
Figure 1.Maximum likelihood tree of Octocorallia based on combined, partitioned analysis of mtMutS, COI and 28S rDNA sequences. Taxa belonging to family Alcyoniidae are shown in blue; new combinations proposed herein are shown in red. Solid circles at nodes indicate strong support from both maximum likelihood (bootstrap values >70%) and Bayesian inference (posterior probability > 0.95); open circles indicate moderate support (bootstrap values >50%, Bayesian pp > 0.95). Strongly supported clades that include no alcyoniid taxa have been collapsed to triangles to facilitate readability. HA: Holaxonia–Alcyoniina clade; CP: Calcaxonia–Pennatulacea clade. Hexacorallian taxa used as outgroups are not shown. For a list of all reference taxa and sequences included in the analysis see Appendix.
-
Figure 10.Two specimens of Cycloseris boschmai sp. n. (RMNH Coel. 8286) with marginal buds and sand in the mouths. Indonesia, Banda, off Lontor, Danish Exp. to the Kei Islands, 15 June 1922.
-
Anna Halász, Catherine S. McFadden, Dafna Aharonovich, Robert Toonen, Yehuda Benayahu
Zookeys
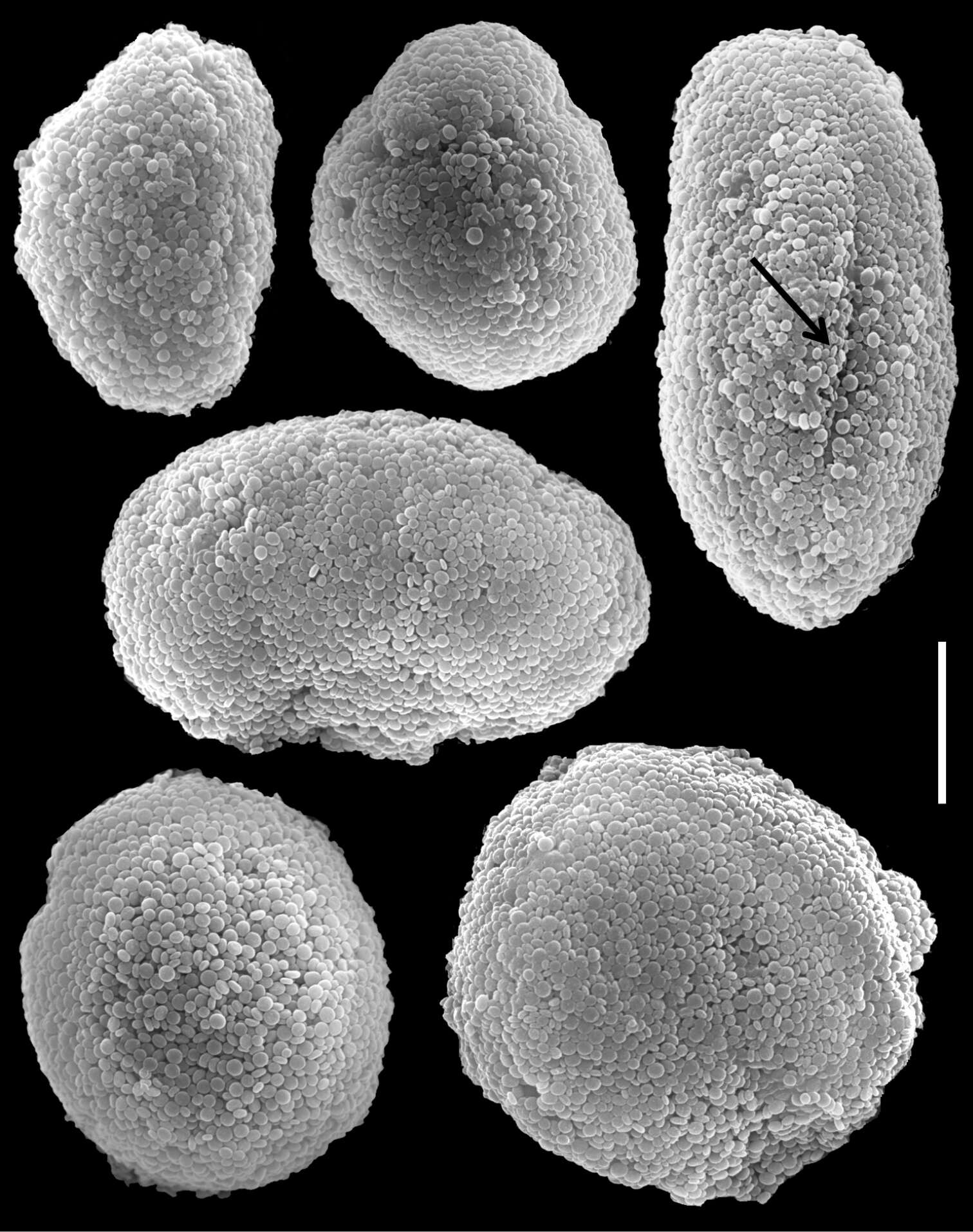
Figure 16.Scanning electron micrographs of polyp sclerites of Ovabunda aldersladei Janes, 2008 holotype (RMNH Coel. 38681). Arrow indicates surface crest. Scale bar 10 µm.
-
Tullia I. Terraneo, Michael L. Berumen, Roberto Arrigoni, Zarinah Waheed, Jessica Bouwmeester, Annalisa Caragnano, Fabrizio Stefani, Francesca Benzoni
Zookeys
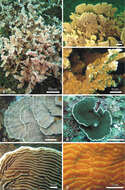
Figure 1.Colonies of Pachyseris rugosa (a–c) and Pachyseris speciosa (d–g) in situ. a Image of the whole colony of specimen IRD HS2893, Prony Bay, New Caledonia b Lateral view of the fronds of specimen IRD HS2594, Prony Bay, New Caledonia c Fronds of specimen IRD HS2856 viewed from above, Prony Bay, New Caledonia d Tiers of foliose projections of a colony from New Caledonia e Image of specimen UNIMIB SO040, Socotra Island f Part of specimen KAUST SA714, Saudi Arabia g Detail of a colony with reduced carinae and brightly colored polyp mouths, growing in very turbid environment, Banc des Japonais, New Caledonia.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.