-
B.A.R. Azman, B.H.R. Othman
Zookeys
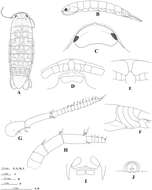
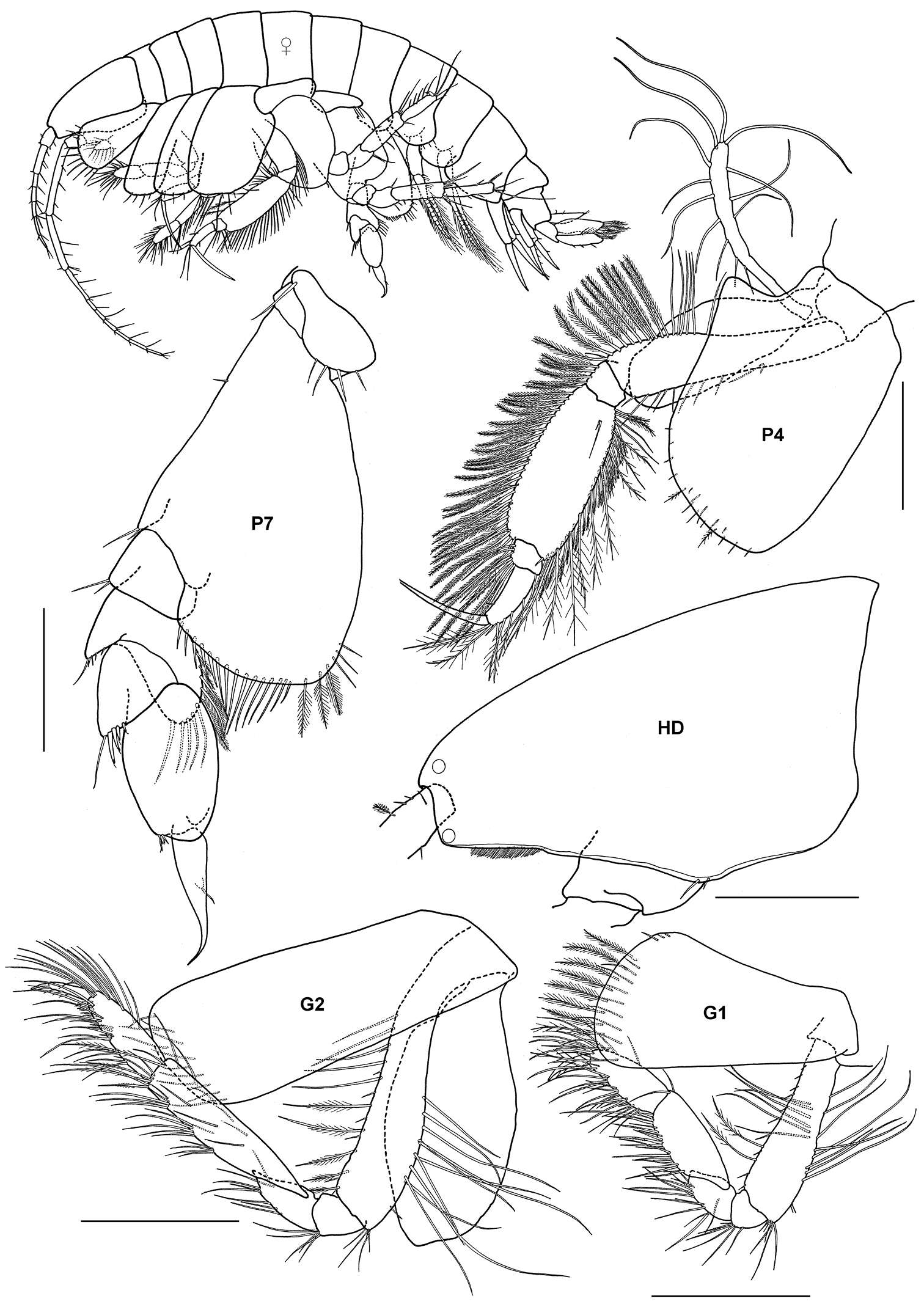
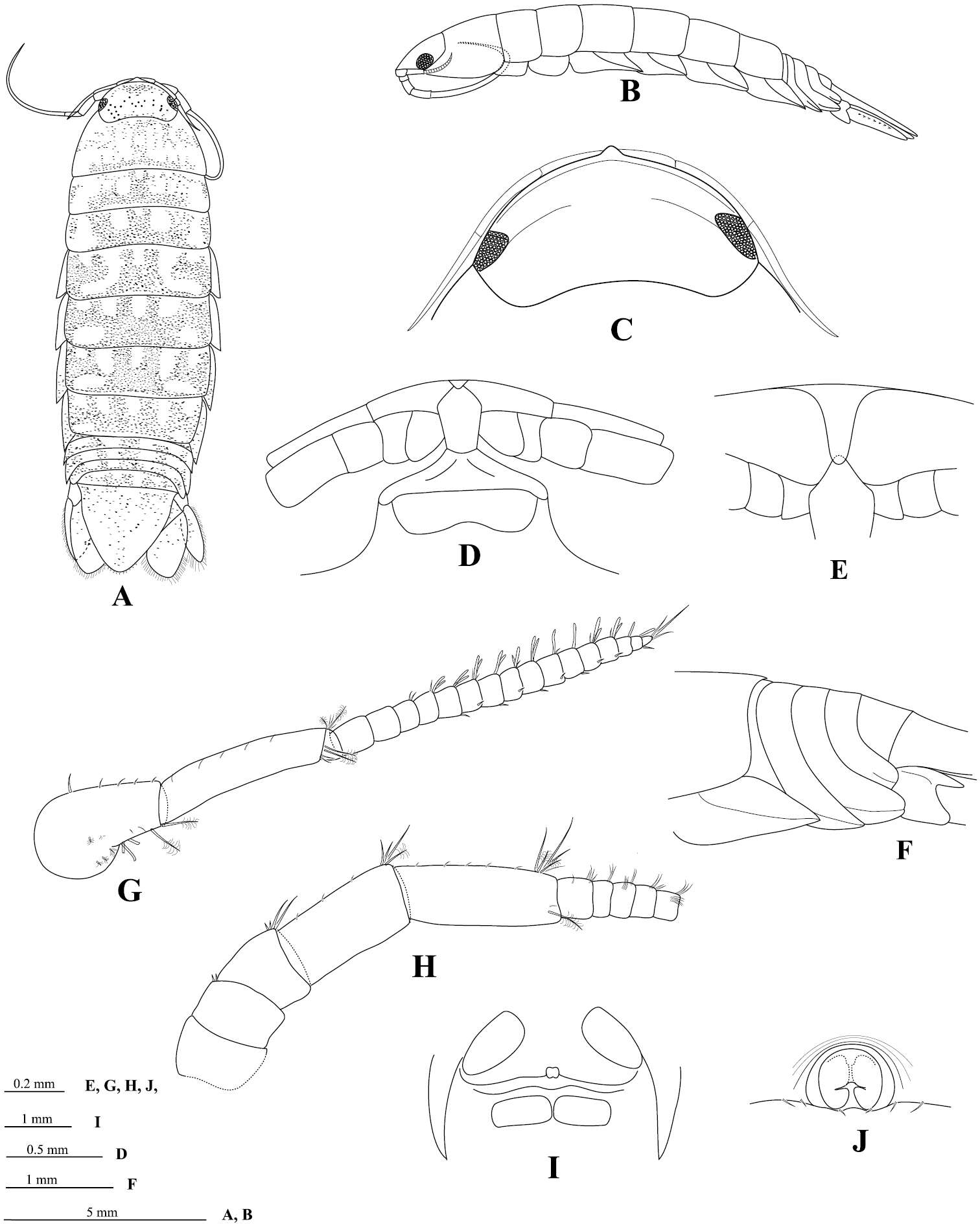
Figure 2.Ampelisca brevicornis (Costa), female (UKMMZ-1454), 4.8 mm. Renggis, Pulau Tioman. Scales for G1, G2, P4, P7 represent 0.5 mm; HD scale = 0.2 mm.
-
Young-Hyo Kim, Ed A. Hendrycks
Zookeys
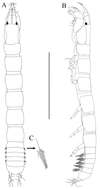
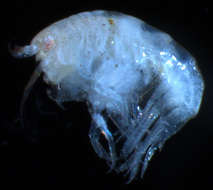
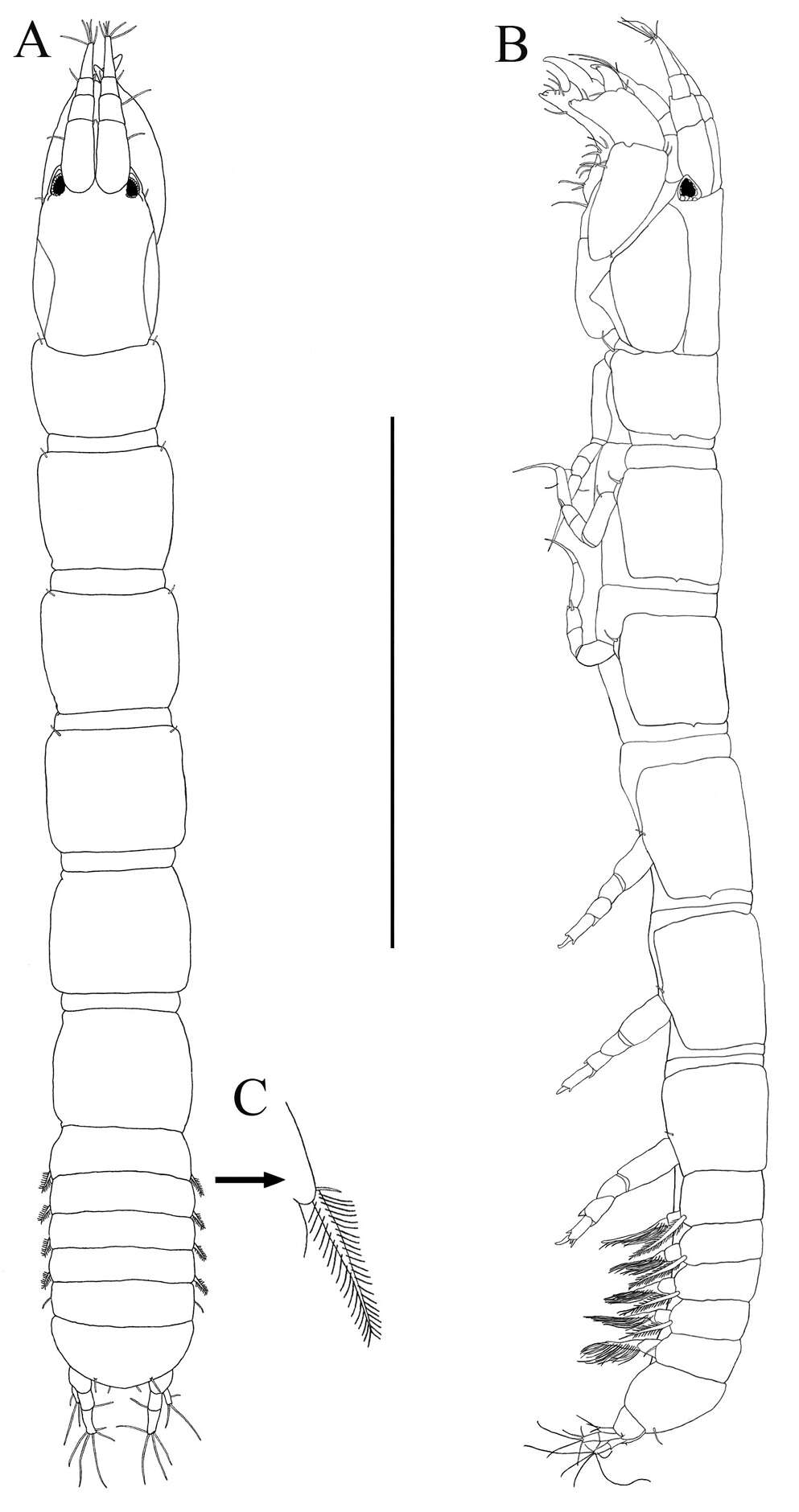
Figure 1.Socarnes tongyeongensis sp. n., female, 8.8 mm, Gyeongpo, Pungwha-ri, Sanyang-eup, Tongyeong-si, Korea.
-
Eknarin Rodcharoen, Niel L. Bruce, Pornsilp Pholpunthin
Zookeys
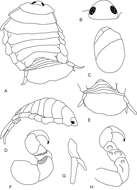
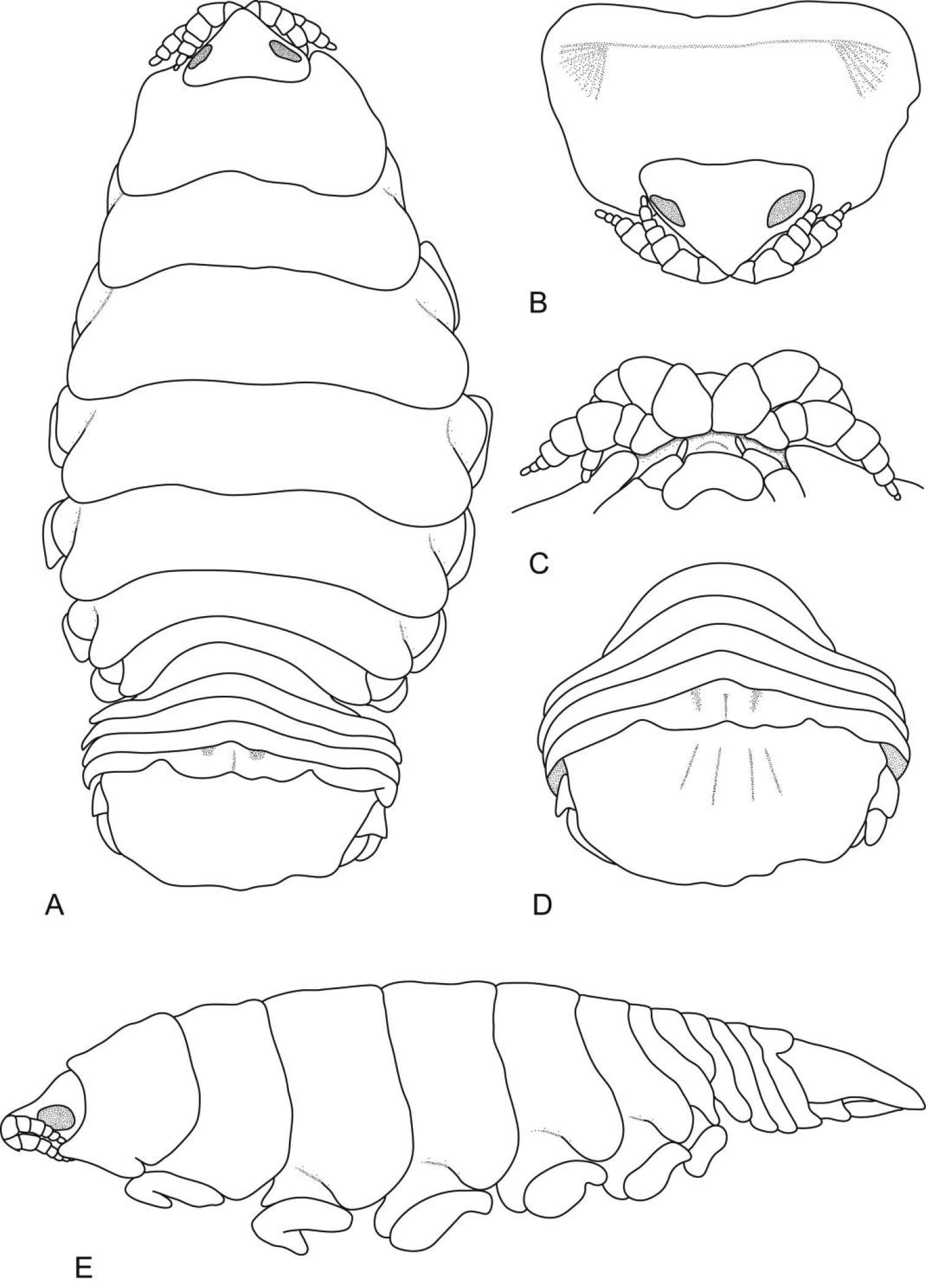
Figure 2.Cirolana songkhla sp. n., male holotype (PSUZC-CR0281-01) (13.7 mm) (A–F), male paratype (PSUZC-CR0281-2) (11.2 mm) (G–H), male paratype (PSUZC-CR0281-2) (13.8 mm) (I–J). A dorsal view B lateral view C head, dorsal view D frons E detail of frontal lamina F pleon G antennule H antennal peduncle I antero-ventral view of penial opening J ventral view of penial opening.
-
Shigenori Karasawa, Kenshi Goto
Zookeys
Figure 1.Burmoniscus kitadaitoensis, male, holotype, TOYA-Cr 14899. A, B Pleopod 1 endopodite C pleopod 1 exopodite D pleopod 2 endopodite E pleopod 2 exopodite F genital papilla. Scale bars: A, C–E 200 μm, B 50 μm.
-
Andrés G. Morales-Núñez, Richard W. Heard
Zookeys
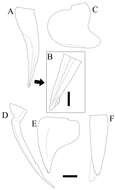
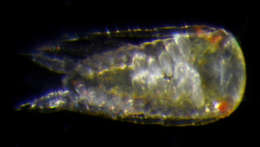
Figure 2.Paratanais rosadi sp. n., holotype female: A dorsal view B lateral view C enlargement of articulated setulate seta on pleonite-1. Scale bar A–B 1.0 mm.
-
Kerry A. Hadfield, Niel L. Bruce, Nico J. Smit
Zookeys
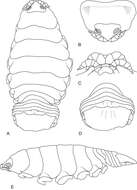
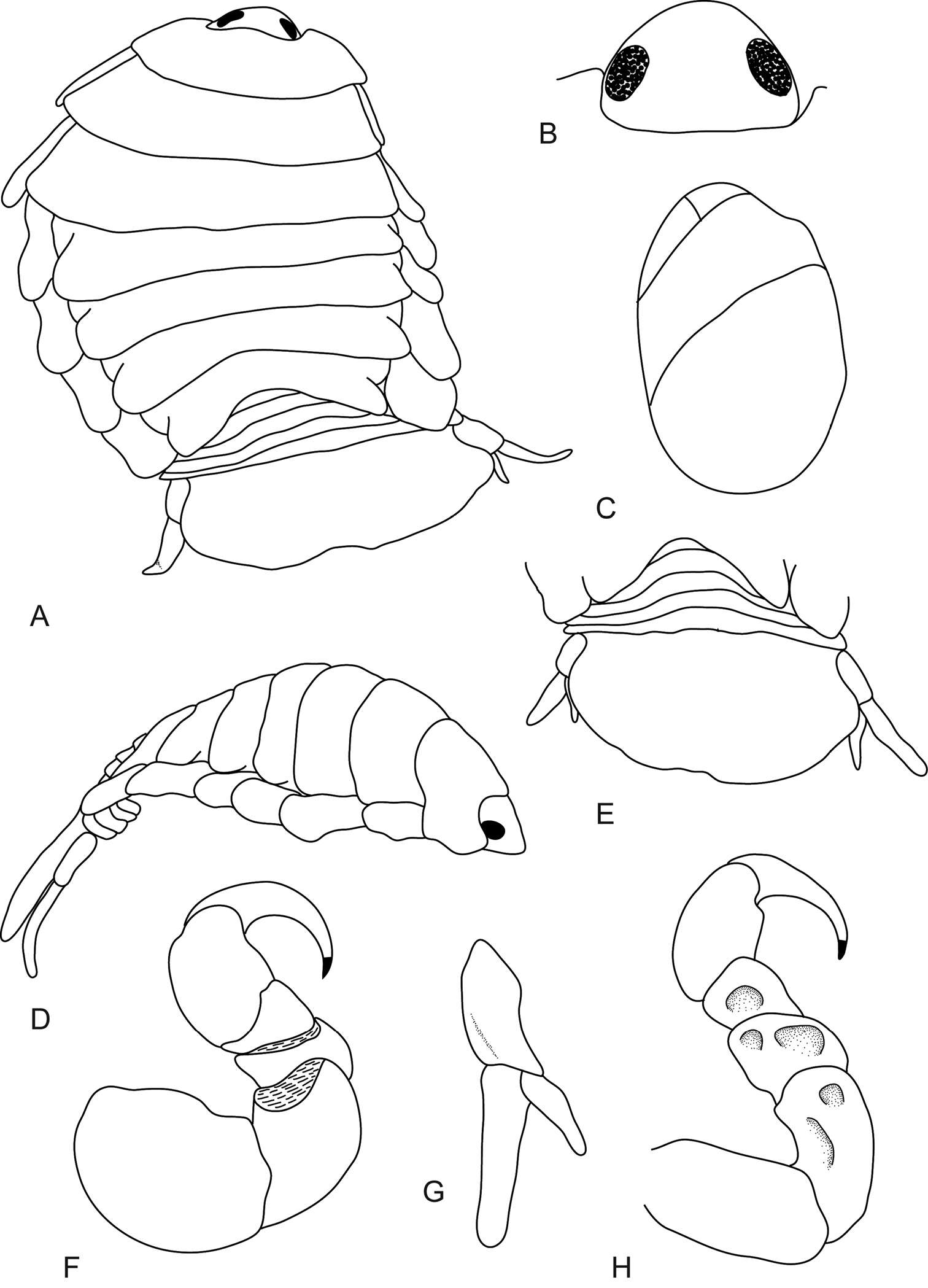
Figure 1.Ceratothoa africanae sp. n. female holotype (29 mm) (SAM A45937): A dorsal view B antero-dorsal view of pereonite 1 and cephalon C ventral view of cephalon D dorsal view of pleotelson E lateral view.
-
Kerry A. Hadfield, Paul C. Sikkel, Nico J. Smit
Zookeys
Figure 1.Mothocya xenobranchia Bruce, 1986 (15 mm) (AMNH_IZC 00197448): A dorsal view B dorsal view of cephalon C oostegites D lateral view E dorsal view of pleotelson F pereopod 1 G uropod H pereopod 7 showing indentations.
-
Kent Island, Chesapeake Bay, Maryland, USA
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.