-
Kristine N. White, James Davis Reimer
Zookeys
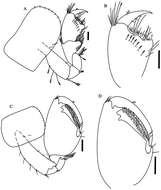
Figure 7.Leucothoe nathani sp. n., holotype male, 4.8 mm, RUMF-ZC-1663.
-
Stella Gomes Rodrigues, Alessandra Angélica de Pádua Bueno, Rodrigo Lopes Ferreira
Zookeys
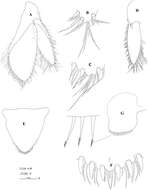
Figure 2.Hyalella imbya sp. n. Rodrigues and Bueno (male paratype, UFLA 0187). A gnathopod 1 B gnathopod 1 propodus and dactylus C gnathopod 2 D gnathopod 2 propodus and dactylus. Scale bars equals 100 µm for A–D.
-
B.A.R. Azman, B.H.R. Othman
Zookeys
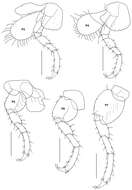
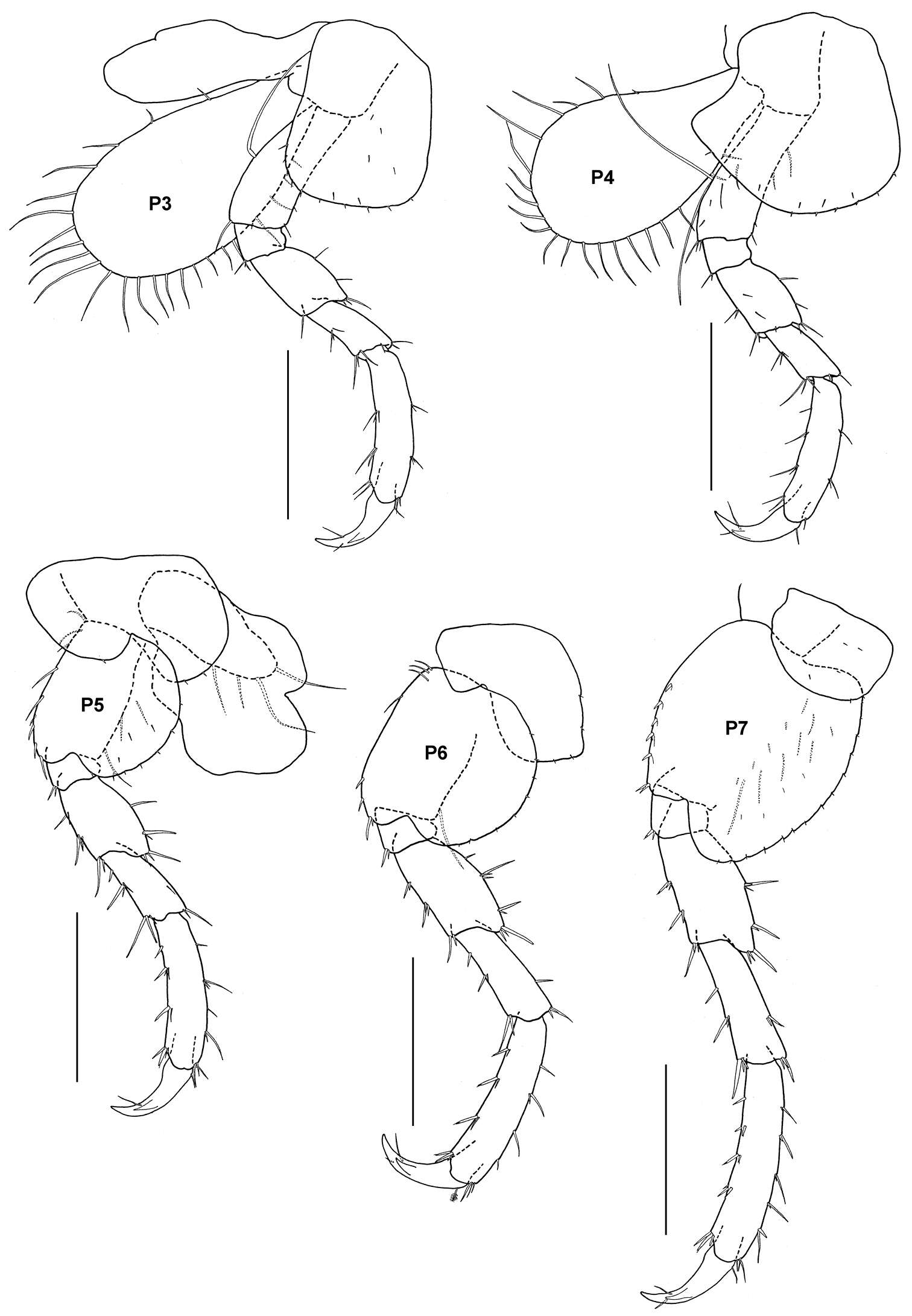
Figure 14.Tethygeneia sunda sp. n., holotype, male (UKMMZ-1252), 4.5 mm. Marine Park, Pulau Tioman. Scales for P3–P7 represent 0.5 mm.
-
Eknarin Rodcharoen, Niel L. Bruce, Pornsilp Pholpunthin
Zookeys
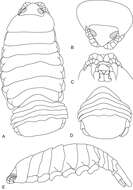
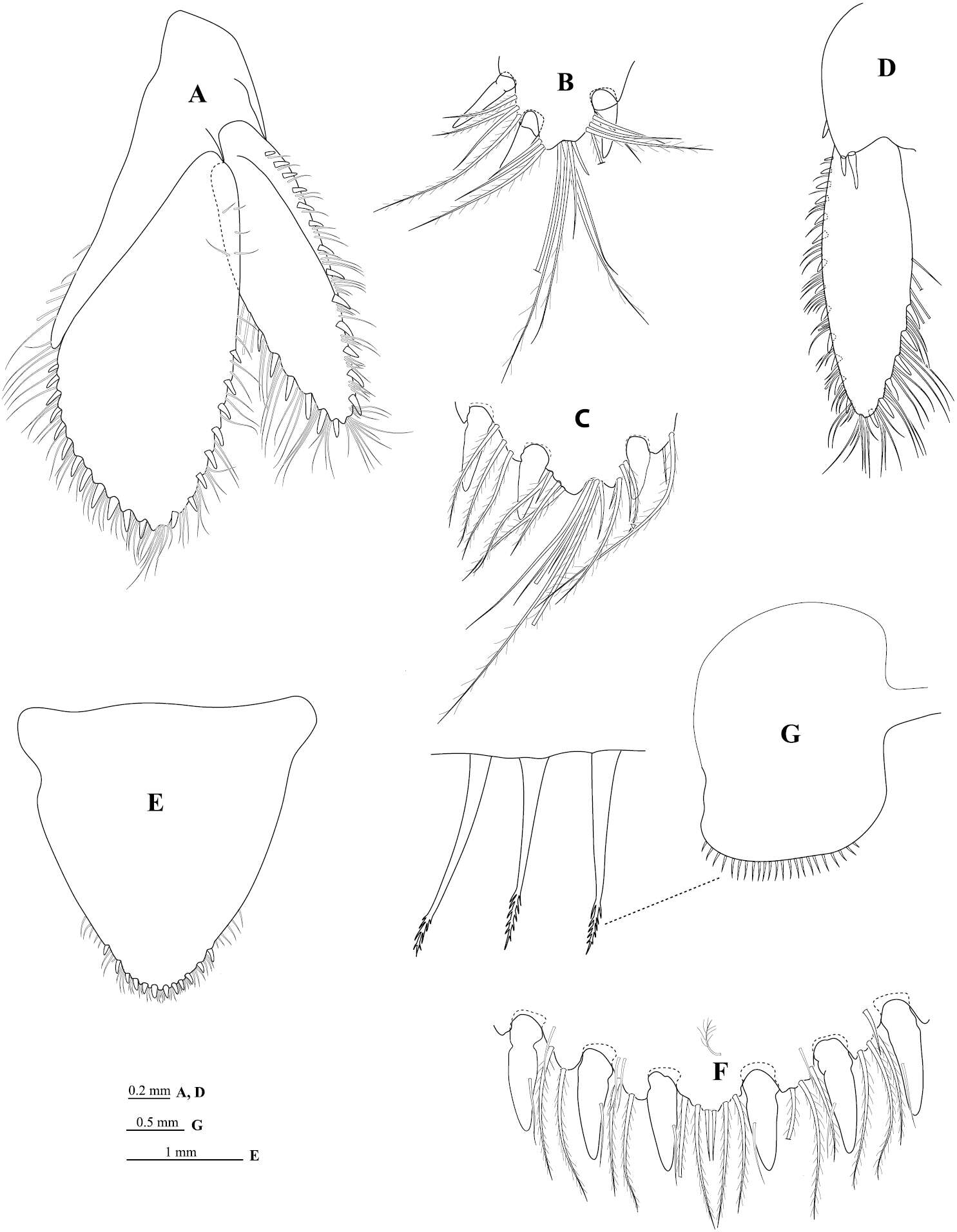
Figure 6.Cirolana songkhla sp. n., male paratype (PSUZC-CR0281-2) (11.2 mm) (A–F), ovigerous female (PSUZC-CR0281-2) (8.7 mm) (G). A uropod B uropod exopod apex C uropod endopod apex D uropod exopod E pleotelson F detail of pleotelson apex G oostegite 4.
-
Andrés G. Morales-Núñez, Richard W. Heard
Zookeys
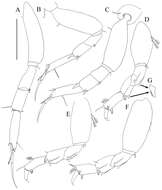
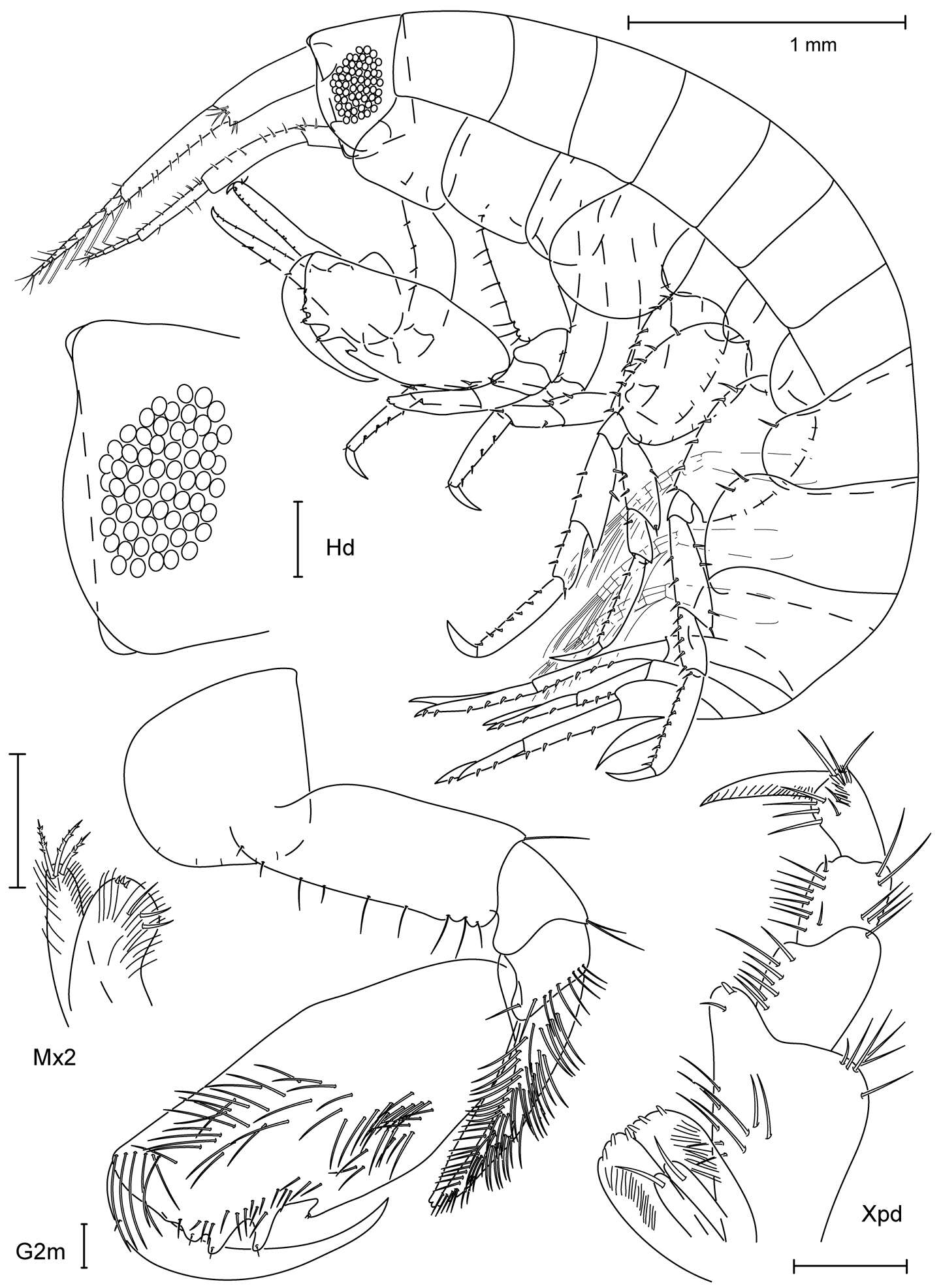
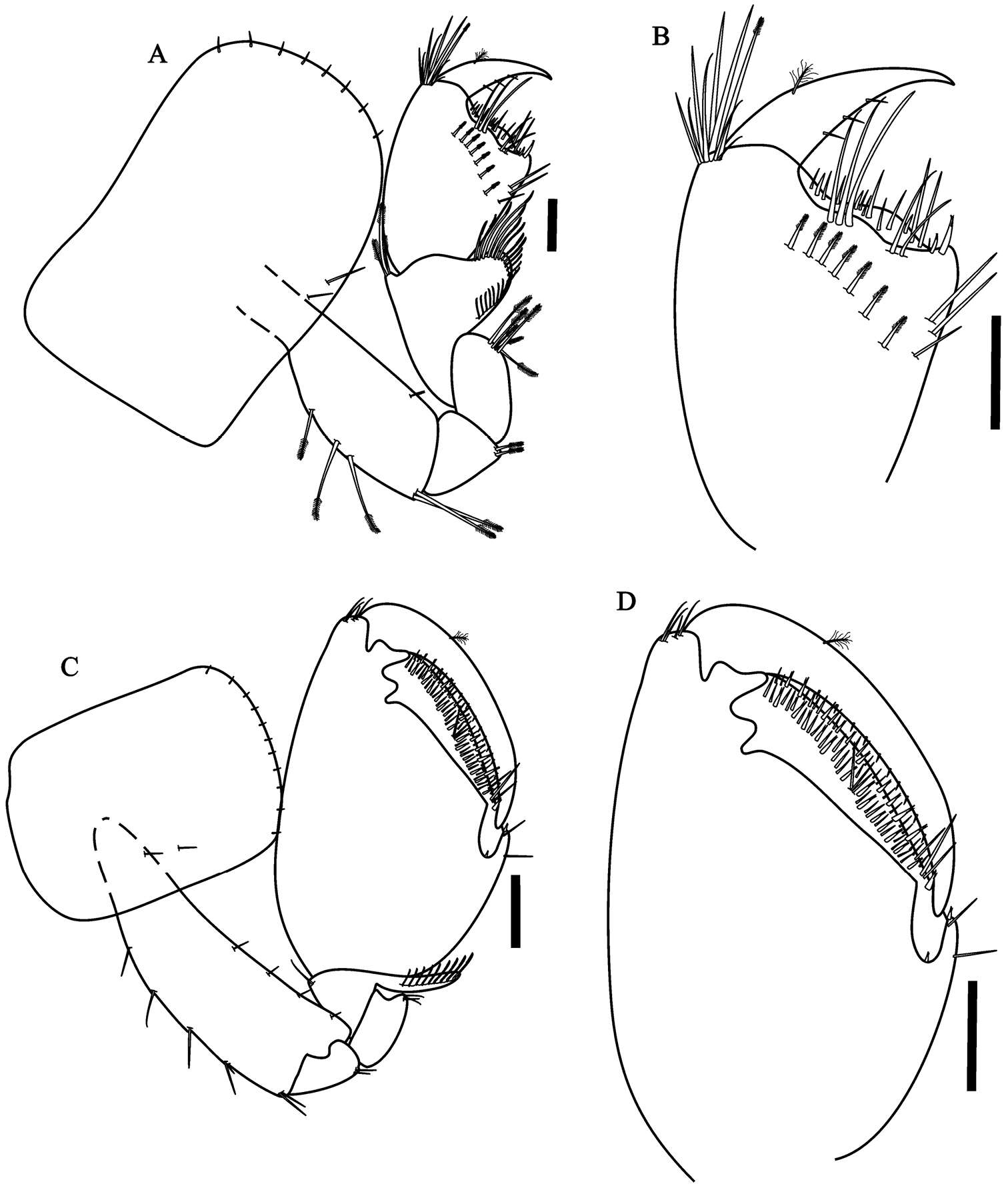
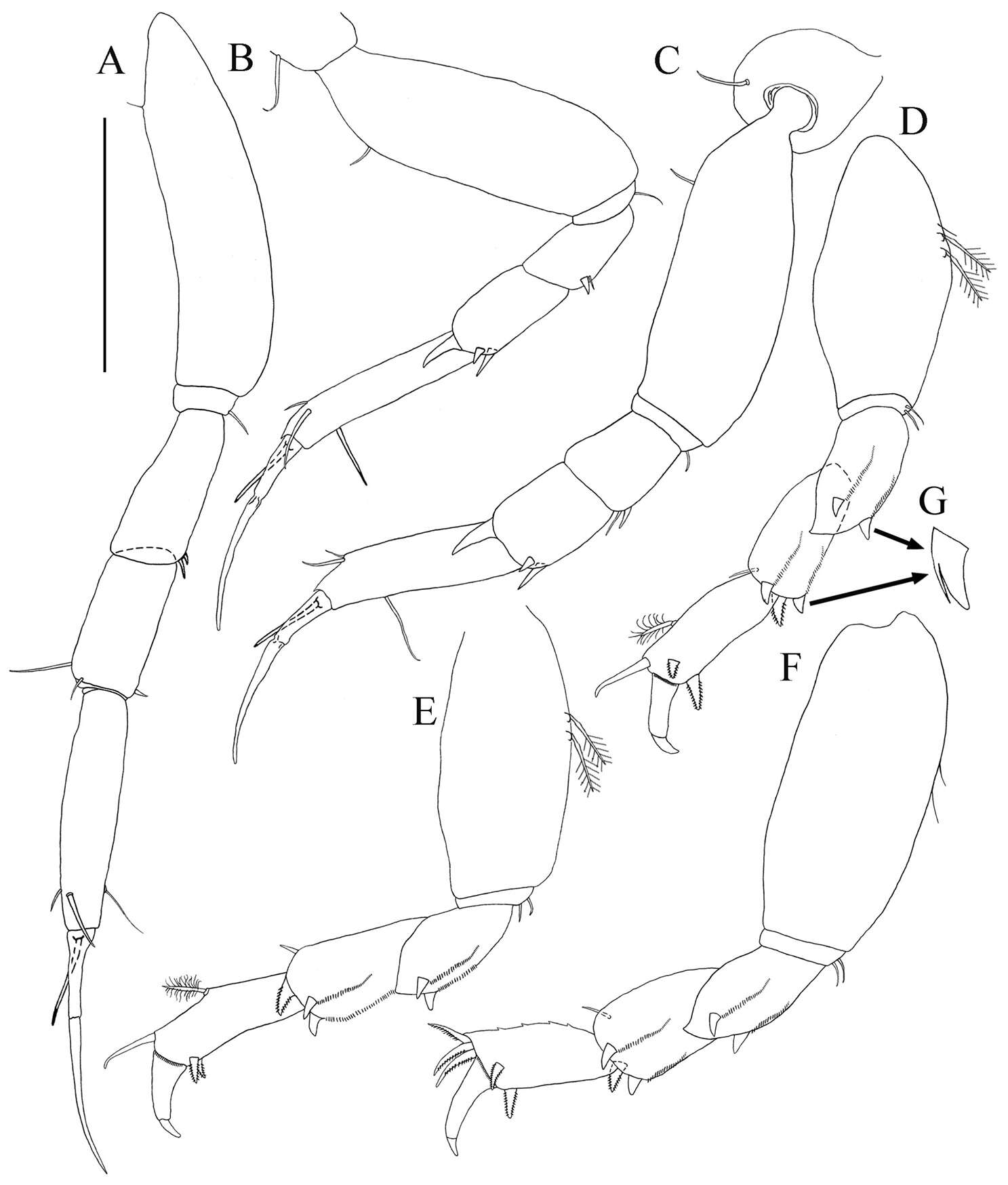
Figure 6.Paratanais rosadi sp. n., holotype female: A pereopod 1 B pereopod 2 C pereopod 3 D pereopod 4 E pereopod 5 F pereopod 6 G enlargement of bifurcate spiniform setae. Scale bar A–F 0.1 mm.
-
Kerry A. Hadfield, Niel L. Bruce, Nico J. Smit
Zookeys
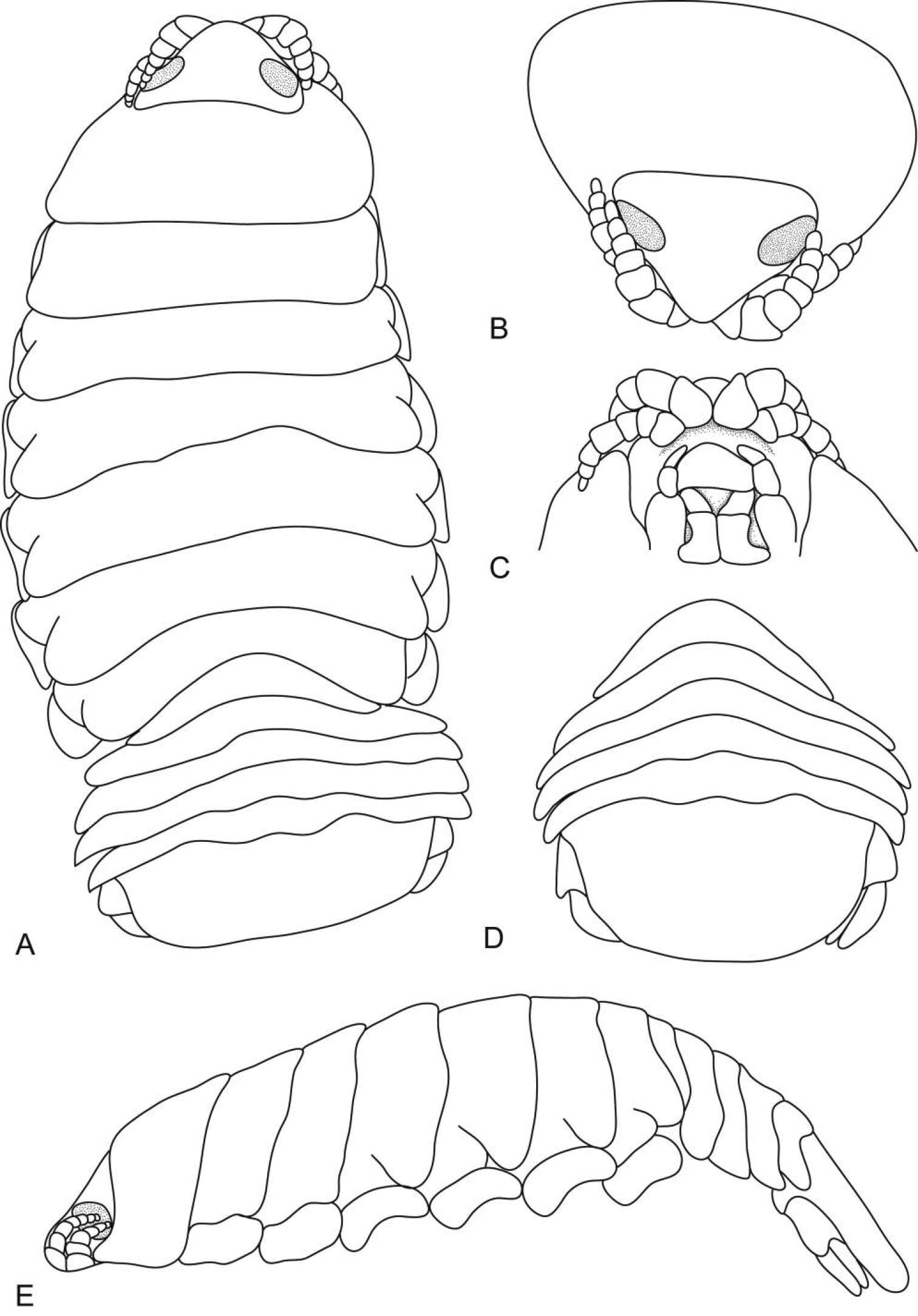
Figure 5.Ceratothoa africanae sp. n. male paratype (14 mm) (SAM A45938): A dorsal view B antero-dorsal view of pereonite 1 and cephalon C ventral view of cephalon D dorsal view of pleotelson E lateral view.
-
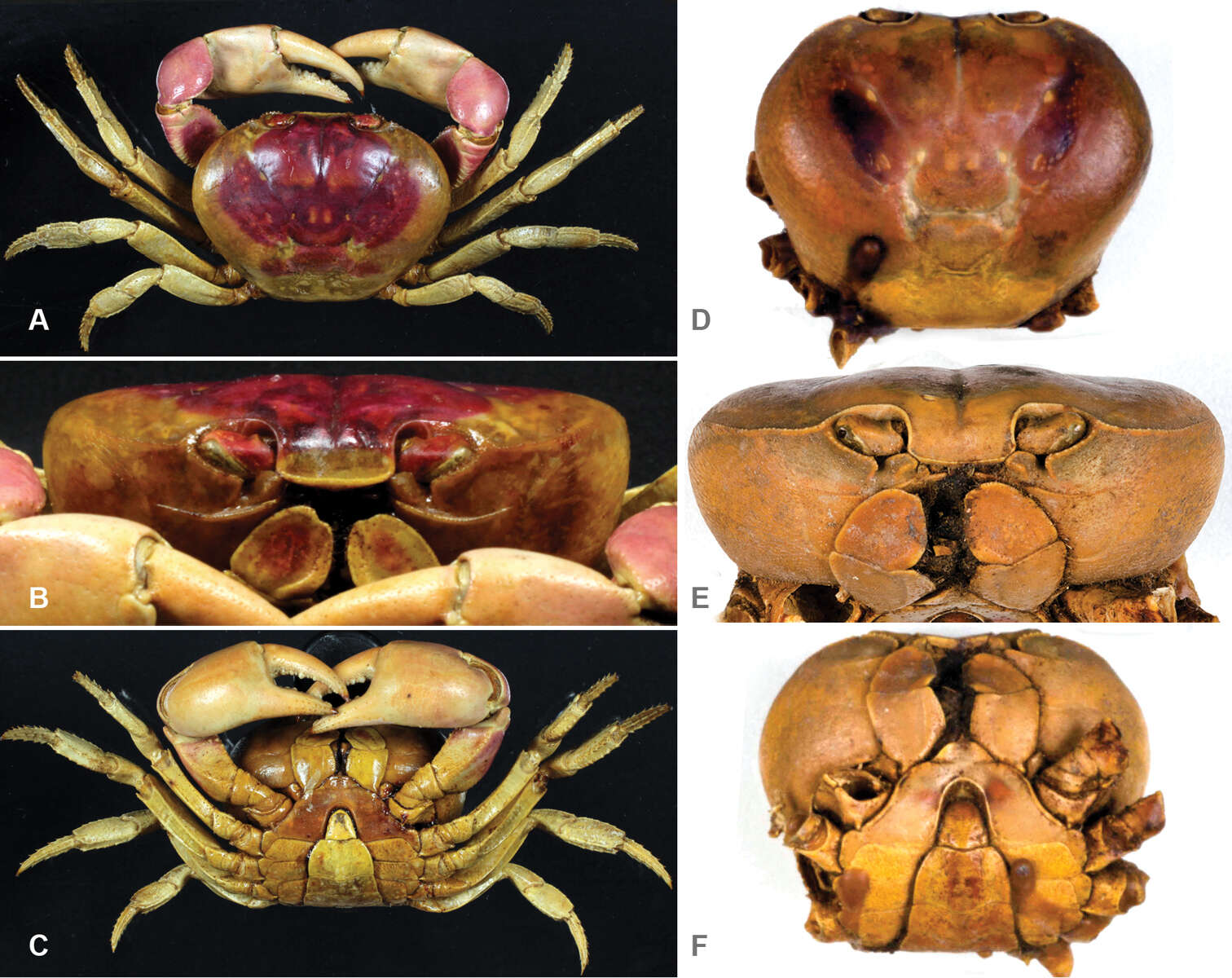
Figure 6.A–C lectotype of Gecarcinus lateralis (Freminville, 1835), male, carapace width 47.2 mm, Guadeloupe (MNHN-3758) (dried specimen, color faded) D–F syntype of Gecarcinus quadratus Saussure, 1853, male, carapace width 39.7 mm, Mexico, Mazatlan (ANSP-CA3741) (dried specimen, color faded) (photos by Paul Callomon, Academy of Natural Sciences of Drexel University, Philadelphia).
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.