-
Caprella spinovissima, Norman.
-
Ampelisca compressa. Near Woods Hole.
-
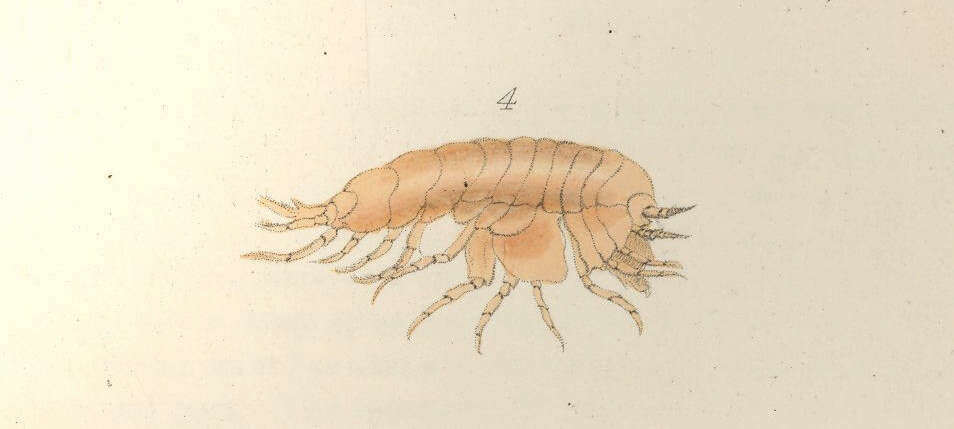
Gammarus lythlops.
-
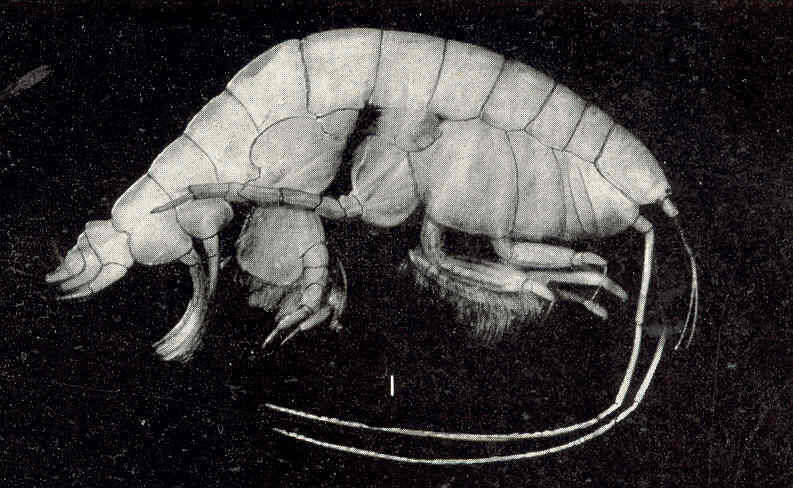
Gammarus locusta. Near Woods Hole.
-
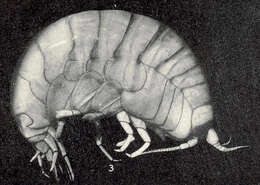
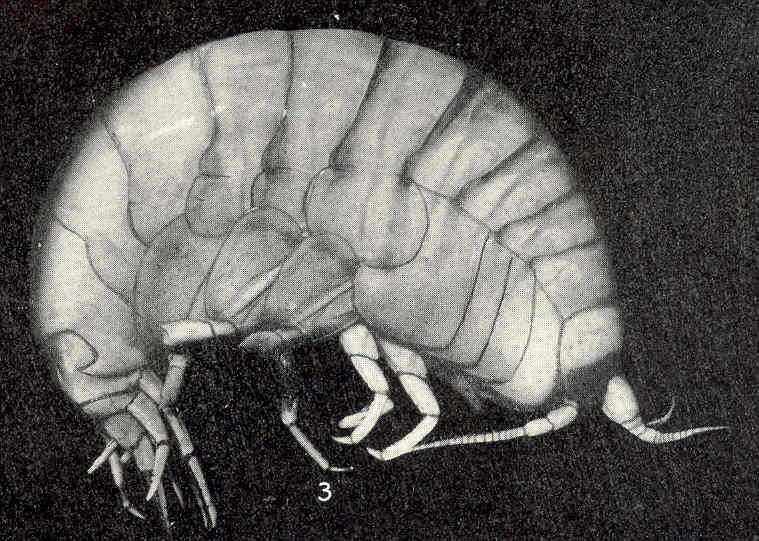
Anonyx nugax. Vineyard Sound.
-
Epimeria loricata. Off Head Harbor, Maine.
-
Ronald Vonk, Bert W. Hoeksema, Damia Jaume
Zookeys
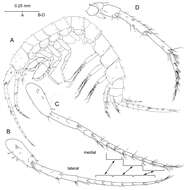
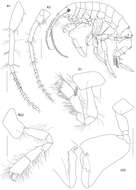
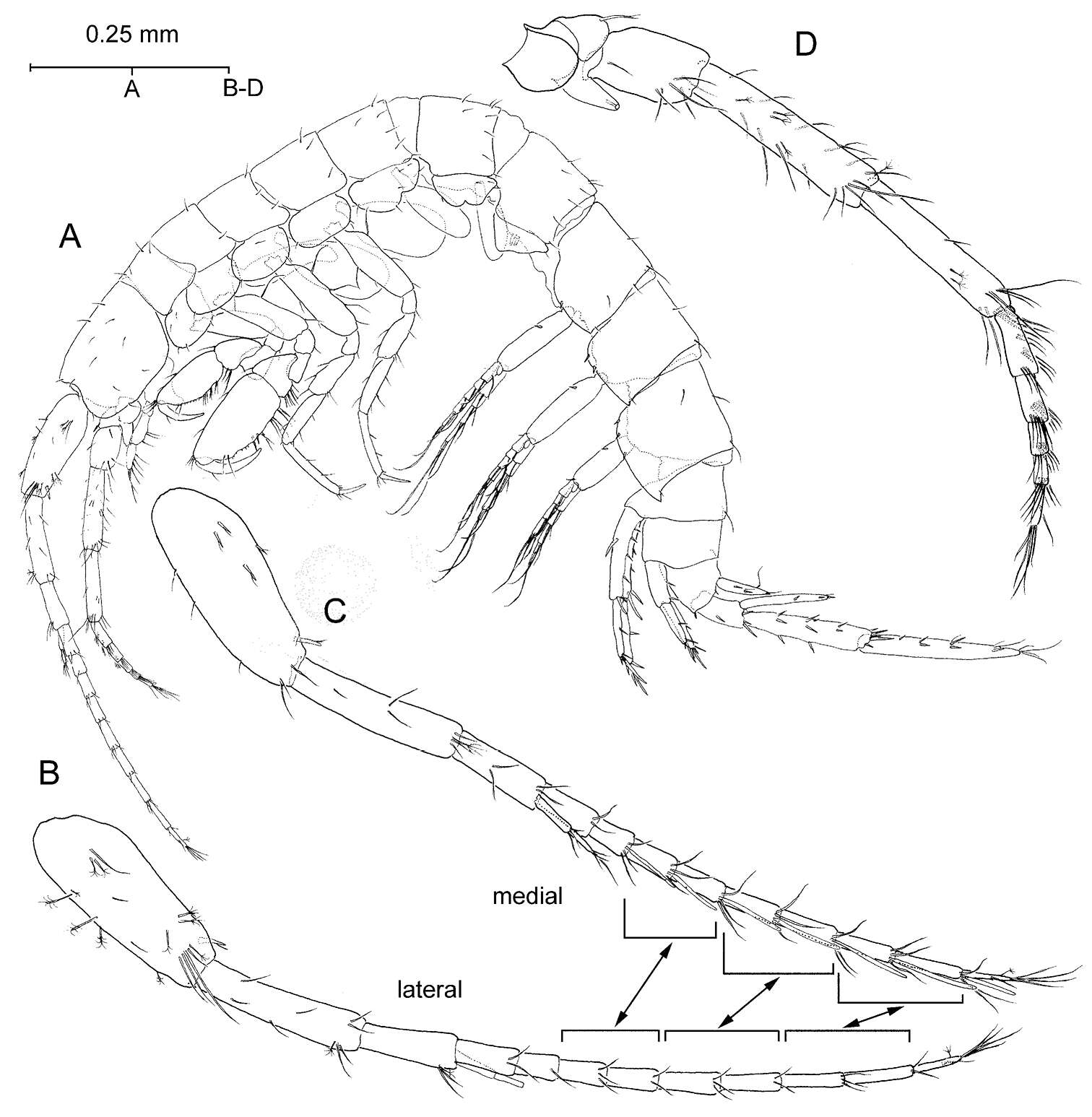
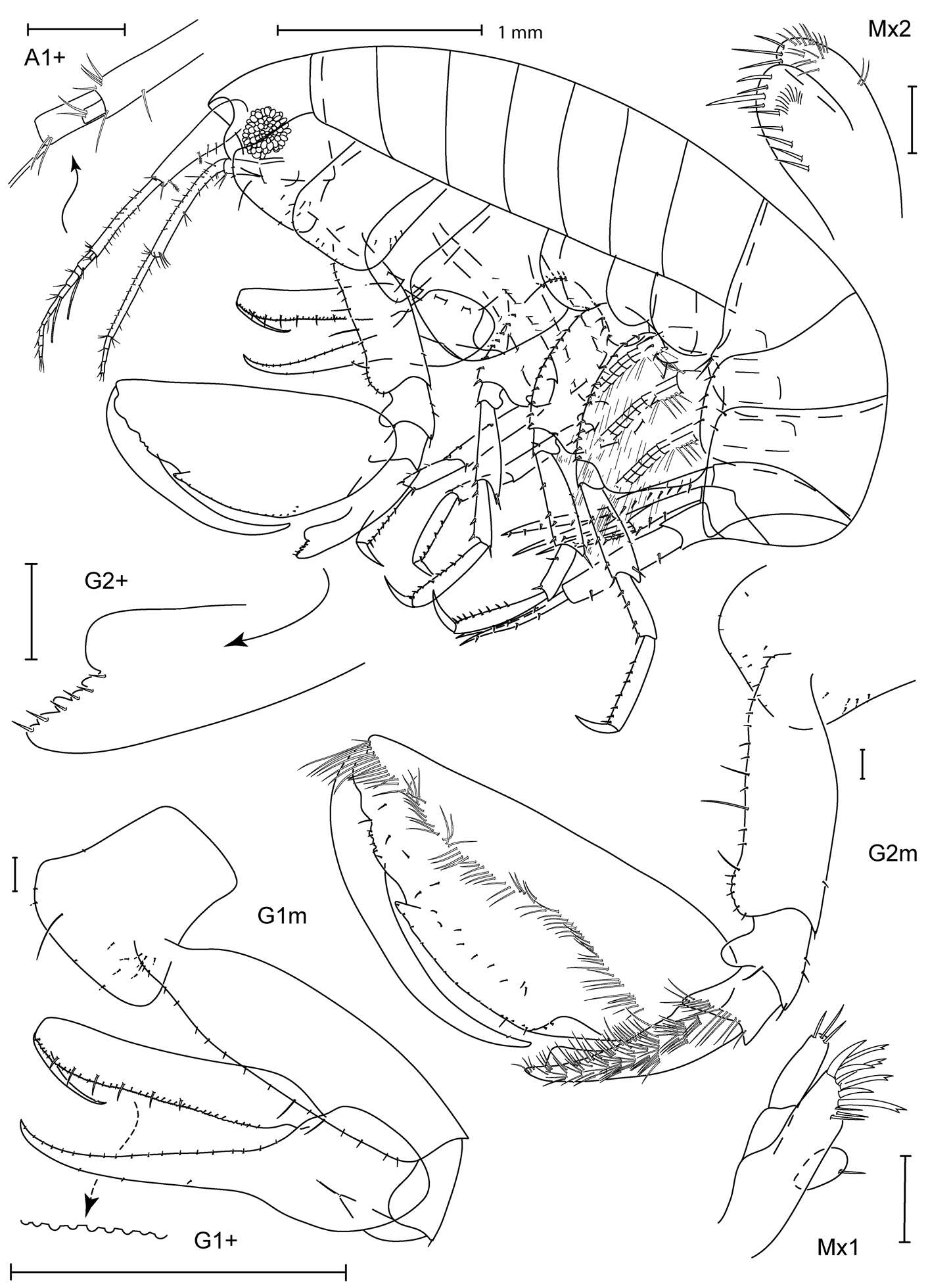
Figure 2.Psammogammarus wallacei sp. n., male paratype 2.53 mm, P5-P7 wanting. A body, lateral B left antennule, lateral C same showing medial armature D right antenna, lateral. Arrows pointing at serially-homologous pairs of articles of main flagellum of antennule.
-
Kristine N. White, James Davis Reimer
Zookeys
Figure 3.Leucothoe amamiensis sp. n., holotype male, 5.9 mm, RUMF-ZC-1654.
-
Stella Gomes Rodrigues, Alessandra Angélica de Pádua Bueno, Rodrigo Lopes Ferreira
Zookeys
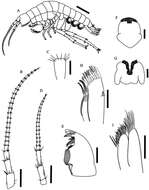
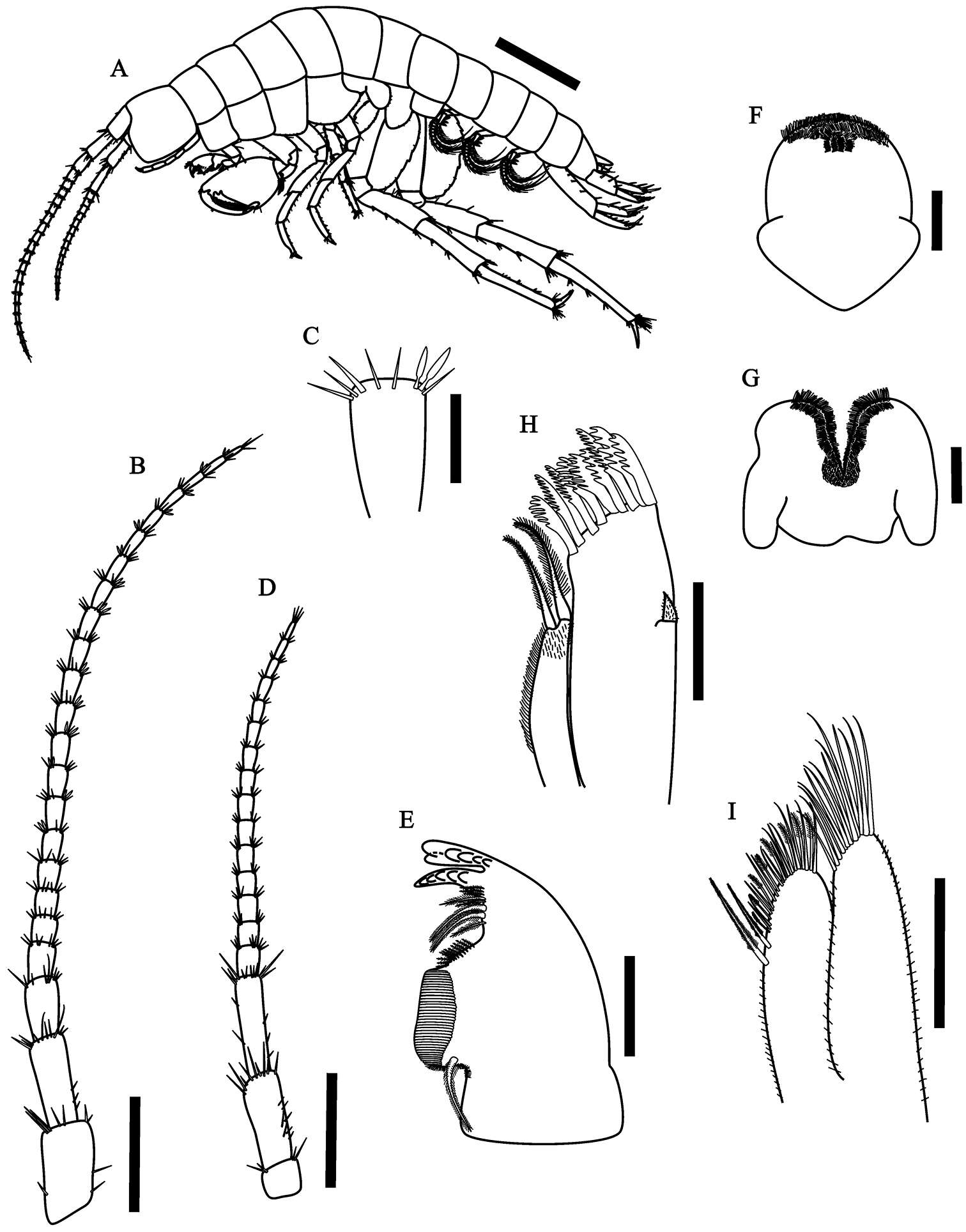
Figure 1.Hyalella imbya sp. n. Rodrigues and Bueno (male paratype, UFLA 0187). A habitus from holotype B antenna 1 C antenna 1 article showing two aesthetascs D antenna 2 E left mandible F upper lip G lower lip H maxilla 1 I maxilla 2. Scale bar equal 1 mm for A; 500 µm for B; 100 µm for C–I.
-
B.A.R. Azman, B.H.R. Othman
Zookeys
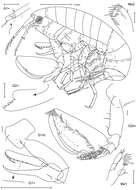
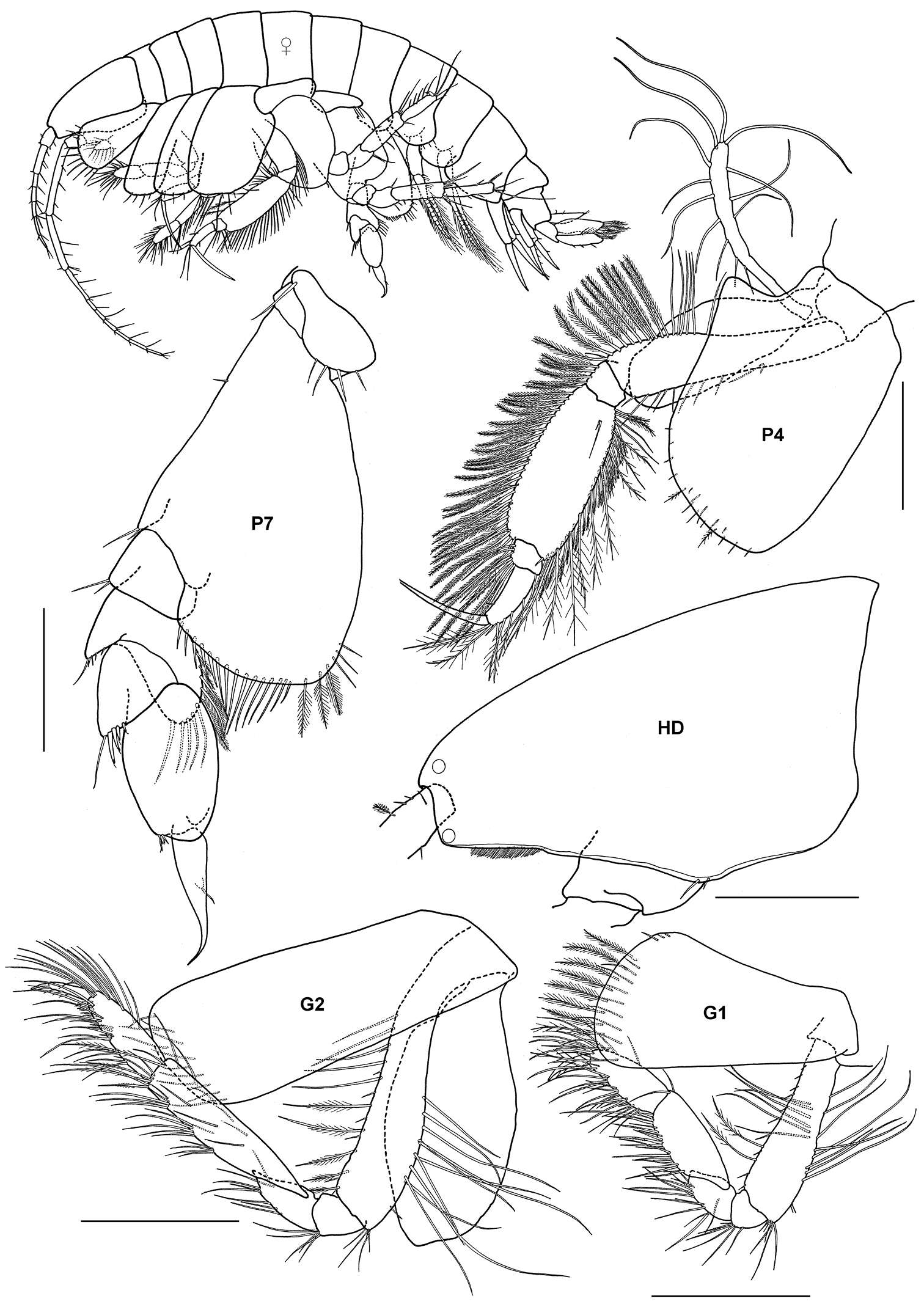
Figure 2.Ampelisca brevicornis (Costa), female (UKMMZ-1454), 4.8 mm. Renggis, Pulau Tioman. Scales for G1, G2, P4, P7 represent 0.5 mm; HD scale = 0.2 mm.
-
Young-Hyo Kim, Ed A. Hendrycks
Zookeys
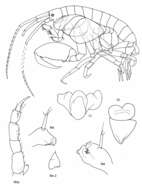
Figure 1.Socarnes tongyeongensis sp. n., female, 8.8 mm, Gyeongpo, Pungwha-ri, Sanyang-eup, Tongyeong-si, Korea.
-
Koraon Wongkamhaeng, Manasawan Saengsakda Pattaratumrong, Ratchanee Puttapreecha
Zookeys
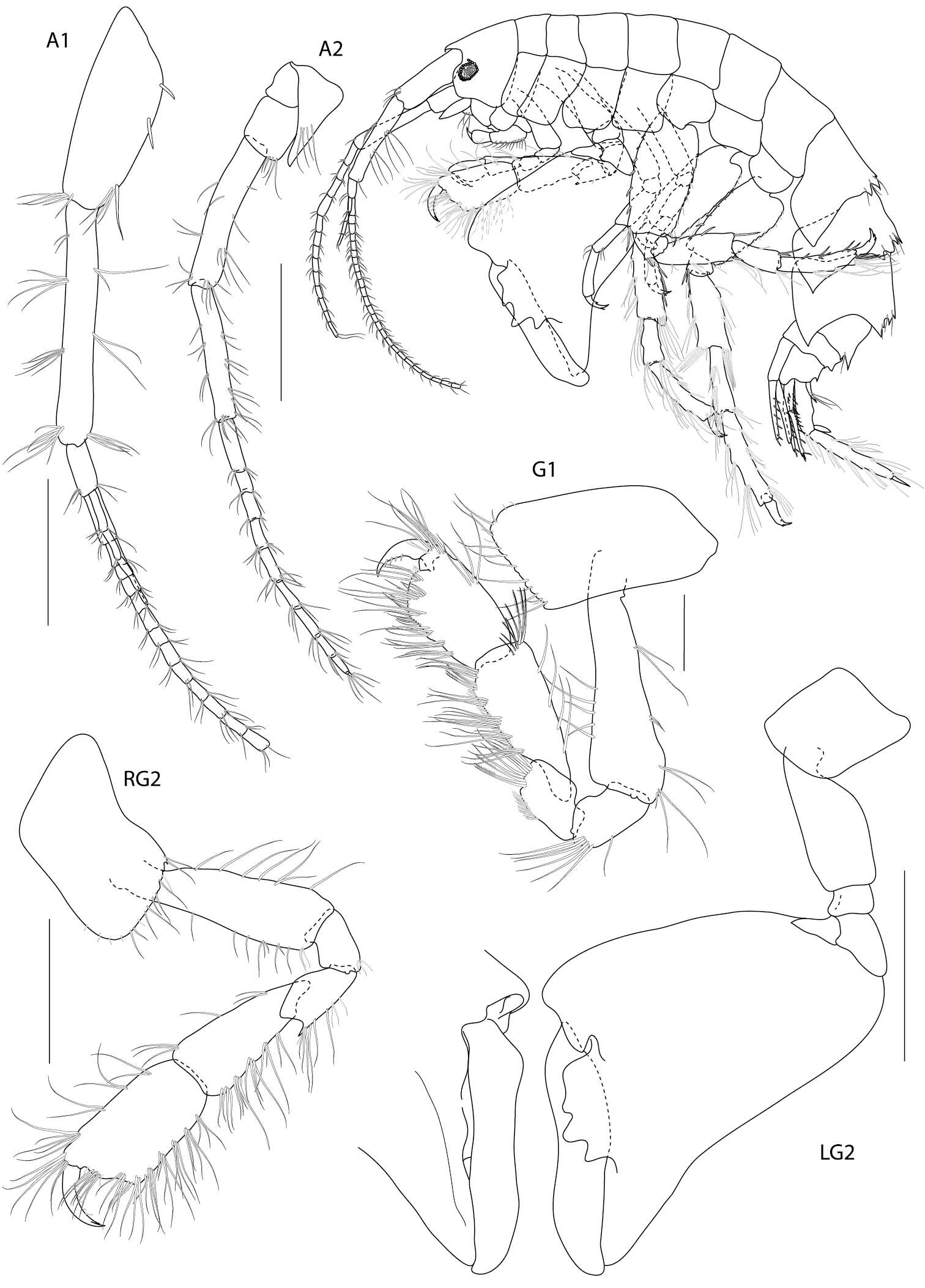
Figure 2.Dulichiella pattaniensis sp. n. holotype, male, (PSUZC-CR-00192), 6.3 mm. Pattani Bay, Lower Gulf of Thailand. All scale bars represent 0.5 mm.
-
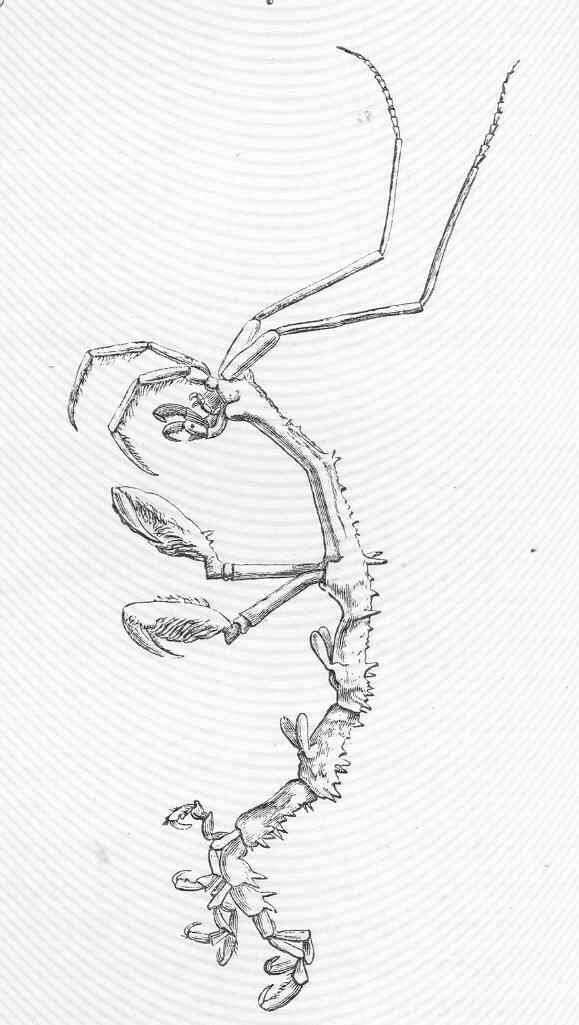
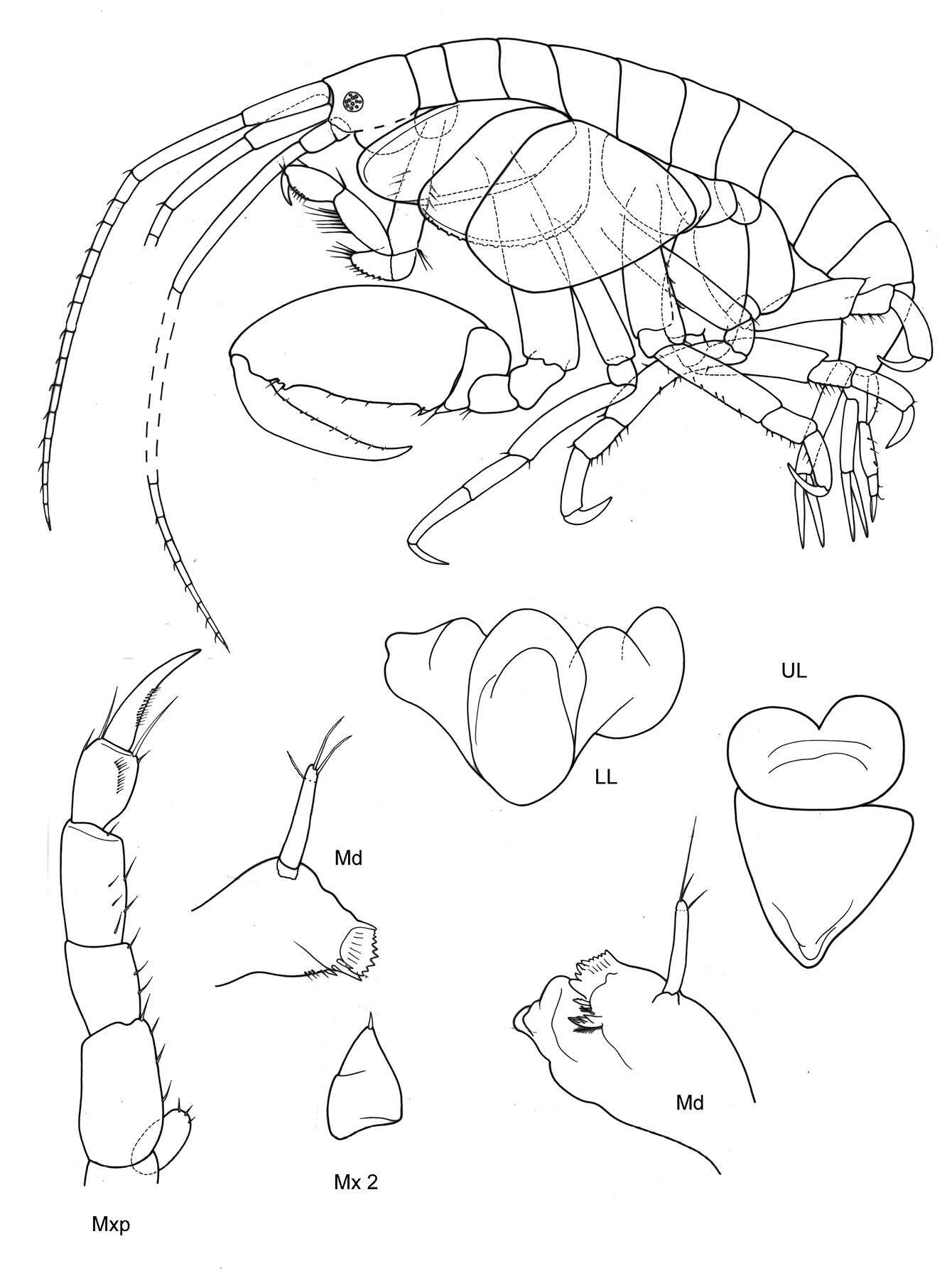
Figure 11.Victometopa rorida gen. n. sp. n.: Habitus ? male 4.4 mm; mouthparts UL, Mx1, 2 Md, LL, Mxp.
-
Kent Island, Chesapeake Bay, Maryland, USA
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.