en
names in breadcrumbs

Abstract:
Klebsiella pneumoniae (K. pneumoniae) is a common nosocomial pathogen causing respiratory tract (pneumoniae) and blood stream infections. Multidrug-resistant (MDR) isolates of K. pneumoniae infections are difficult to treat in patients in health care settings. Aim of the present study was to determine the impact of Mr. Trivedi’s biofield treatment on four MDR clinical lab isolates (LS) of K. pneumoniae (LS 2, LS 6, LS 7, and LS 14). Samples were divided into two groups i.e. control and biofield treated. Control and treated groups were analyzed for antimicrobial susceptibility pattern, minimum inhibitory concentration (MIC), biochemical study and biotype number using MicroScan Walk-Away® system. The analysis was done on day 10 after biofield treatment as compared with control group. Antimicrobial sensitivity assay showed that there was 46.42% alteration in sensitivity of tested antimicrobials in treated group of MDR K. pneumonia isolates. MIC results showed an alteration in 30% of tested antimicrobials out of thirty after biofield treatment in clinical isolates of K. pneumoniae. An increase in antimicrobial sensitivity and decrease in MIC value was reported (in LS 6) in case of piperacillin/tazobactam and piperacillin. Biochemical study showed a 15.15% change in biochemical reactions as compared to control. A significant change in biotype numbers were reported in all four clinical isolates of MDR K. pneumoniae after biofield treatment as compared to control group. On the basis of changed biotype number after biofield treatment, new organism was identified as Enterobacter aerogenes in LS 2 and LS 14. These results suggest that biofield treatment has a significant effect on altering the antimicrobial sensitivity, MIC values, biochemical reactions and biotype number of multidrug-resistant isolates of K. pneumoniae.
Klebsiella pneumoniae (K. pneumoniae) is a common nosocomial pathogen causing respiratory tract (pneumoniae) and blood stream infections. Multidrug-resistant (MDR) isolates of K. pneumoniae infections are difficult to treat in patients in health care settings. Aim of the present study was to determine the impact of Mr. Trivedi’s biofield treatment on four MDR clinical lab isolates (LS) of K. pneumoniae (LS 2, LS 6, LS 7, and LS 14). Samples were divided into two groups i.e. control and biofield treated. Control and treated groups were analyzed for antimicrobial susceptibility pattern, minimum inhibitory concentration (MIC), biochemical study and biotype number using MicroScan Walk-Away® system. The analysis was done on day 10 after biofield treatment as compared with control group. Antimicrobial sensitivity assay showed that there was 46.42% alteration in sensitivity of tested antimicrobials in treated group of MDR K. pneumonia isolates. MIC results showed an alteration in 30% of tested antimicrobials out of thirty after biofield treatment in clinical isolates of K. pneumoniae. An increase in antimicrobial sensitivity and decrease in MIC value was reported (in LS 6) in case of piperacillin/tazobactam and piperacillin. Biochemical study showed a 15.15% change in biochemical reactions as compared to control. A significant change in biotype numbers were reported in all four clinical isolates of MDR K. pneumoniae after biofield treatment as compared to control group. On the basis of changed biotype number after biofield treatment, new organism was identified as Enterobacter aerogenes in LS 2 and LS 14. These results suggest that biofield treatment has a significant effect on altering the antimicrobial sensitivity, MIC values, biochemical reactions and biotype number of multidrug-resistant isolates of K. pneumoniae.
Klebsiella pneumoniae (K. pneumoniae) is a common nosocomial pathogen causing respiratory tract (pneumoniae) and blood stream infections. Multidrug-resistant (MDR) isolates of K. pneumoniae infections are difficult to treat in patients in health care settings. Aim of the present study was to determine the impact of Mr. Trivedi’s biofield treatment on four MDR clinical lab isolates (LS) of K. pneumoniae (LS 2, LS 6, LS 7, and LS 14). Samples were divided into two groups i.e. control and biofield treated. Control and treated groups were analyzed for antimicrobial susceptibility pattern, minimum inhibitory concentration (MIC), biochemical study and biotype number using MicroScan Walk-Away® system. The analysis was done on day 10 after biofield treatment as compared with control group. Antimicrobial sensitivity assay showed that there was 46.42% alteration in sensitivity of tested antimicrobials in treated group of MDR K. pneumonia isolates. MIC results showed an alteration in 30% of tested antimicrobials out of thirty after biofield treatment in clinical isolates of K. pneumoniae. An increase in antimicrobial sensitivity and decrease in MIC value was reported (in LS 6) in case of piperacillin/tazobactam and piperacillin. Biochemical study showed a 15.15% change in biochemical reactions as compared to control. A significant change in biotype numbers were reported in all four clinical isolates of MDR K. pneumoniae after biofield treatment as compared to control group. On the basis of changed biotype number after biofield treatment, new organism was identified as Enterobacter aerogenes in LS 2 and LS 14. These results suggest that biofield treatment has a significant effect on altering the antimicrobial sensitivity, MIC values, biochemical reactions and biotype number of multidrug-resistant isolates of K. pneumoniae.
Abstract:
Pathogenic isolates of Klebsiella pneumoniae (K. pneumoniae), particularly the extended-spectrum β-lactamase (ESBL) producing strains, are mostly associated with the failure of antibiotic therapy in nosocomial infections. The present work was designed to evaluate the impact of Mr. Trivedi’s biofield energy treatment on phenotypic and genotypic characteristics of K. pneumoniae. The strain of K. pneumoniae bearing ATCC 15380 (American Type Culture Collection) was procured from the Bangalore Genei, in sealed pack and divided into control and treated groups. Treated group was subjected to Mr. Trivedi’s biofield energy treatment and analyzed for the antimicrobial susceptibility, minimum inhibitory concentration (MIC), biochemical reactions, and biotyping using automated MicroScan Walk-Away® system. Further, the effect of biofield treatment was also evaluated using Random Amplified Polymorphic DNA (RAPD) in order to determine their epidemiological relatedness and genetic characteristics of biofield treated K. pneumoniae samples. The antimicrobial susceptibility results showed an improve sensitivity (i.e. from intermediate to susceptible) of ampicillin/sulbactam and chloramphenicol, while altered sensitivity of cephalothin (i.e. from susceptible to intermediate) was also reported as compared to the control sample. The MIC value showed two-fold decrease in MIC value of ampicillin/sulbactam (i.e. 16/8 to ≤8/4 μg/mL) and chloramphenicol (i.e. 16 to ≤ 8 μg/mL) as compared to the control. The cephalothin showed two-folds change (i.e. ≤ 8 to 16 μg/mL) in the MIC value as compared with the control. Biofield treatment showed 9.09% alterations in biochemical reactions followed by a change in biotype number (7774 4272) in the treated group with respect to the control (7774 4274). Genetic fingerprinting was performed on control and treated samples using RAPD-PCR biomarkers, which showed an average range of 11 to 15% of polymorphism among the treated samples with respect to the control. These results suggested that Mr. Trivedi’s biofield energy treatment has a significant impact on K. pneumoniae.
Pathogenic isolates of Klebsiella pneumoniae (K. pneumoniae), particularly the extended-spectrum β-lactamase (ESBL) producing strains, are mostly associated with the failure of antibiotic therapy in nosocomial infections. The present work was designed to evaluate the impact of Mr. Trivedi’s biofield energy treatment on phenotypic and genotypic characteristics of K. pneumoniae. The strain of K. pneumoniae bearing ATCC 15380 (American Type Culture Collection) was procured from the Bangalore Genei, in sealed pack and divided into control and treated groups. Treated group was subjected to Mr. Trivedi’s biofield energy treatment and analyzed for the antimicrobial susceptibility, minimum inhibitory concentration (MIC), biochemical reactions, and biotyping using automated MicroScan Walk-Away®system. Further, the effect of biofield treatment was also evaluated using Random Amplified Polymorphic DNA (RAPD) in order to determine their epidemiological relatedness and genetic characteristics of biofield treated K. pneumoniaesamples. The antimicrobial susceptibility results showed an improve sensitivity (i.e. from intermediate to susceptible) of ampicillin/sulbactam and chloramphenicol, while altered sensitivity of cephalothin (i.e. from susceptible to intermediate) was also reported as compared to the control sample. The MIC value showed two-fold decrease in MIC value of ampicillin/sulbactam (i.e. 16/8 to ≤8/4 μg/mL) and chloramphenicol (i.e. 16 to ≤ 8 μg/mL) as compared to the control. The cephalothin showed two-folds change (i.e. ≤ 8 to 16 μg/mL) in the MIC value as compared with the control. Biofield treatment showed 9.09% alterations in biochemical reactions followed by a change in biotype number (7774 4272) in the treated group with respect to the control (7774 4274). Genetic fingerprinting was performed on control and treated samples using RAPD-PCR biomarkers, which showed an average range of 11 to 15% of polymorphism among the treated samples with respect to the control. These results suggested that Mr. Trivedi’s biofield energy treatment has a significant impact on K. pneumoniae.
Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated, lactose-fermenting, facultative anaerobic, rod-shaped bacterium. It appears as a mucoid lactose fermenter on MacConkey agar.
Although found in the normal flora of the mouth, skin, and intestines,[1] it can cause destructive changes to human and animal lungs if aspirated, specifically to the alveoli resulting in bloody, brownish or yellow colored jelly-like sputum. In the clinical setting, it is the most significant member of the genus Klebsiella of the Enterobacteriaceae. K. oxytoca and K. rhinoscleromatis have also been demonstrated in human clinical specimens. In recent years, Klebsiella species have become important pathogens in nosocomial infections.
It naturally occurs in the soil, and about 30% of strains can fix nitrogen in anaerobic conditions.[2] As a free-living diazotroph, its nitrogen-fixation system has been much-studied, and is of agricultural interest, as K. pneumoniae has been demonstrated to increase crop yields in agricultural conditions.[3]
It is closely related to K. oxytoca from which it is distinguished by being indole-negative and by its ability to grow on melezitose but not 3-hydroxybutyrate.
The genus Klebsiella was named after the German microbiologist Edwin Klebs (1834–1913). It is also known as Friedlander's bacillum in honor of Carl Friedländer, a German pathologist, who proposed that this bacterium was the etiological factor for the pneumonia seen especially in immunocompromised individuals such as people with chronic diseases or alcoholics.
Community-acquired pneumonia caused by Klebsiella pneumoniae may occasionally be called Friedländer's pneumonia.[4]
Illness most commonly affects middle-aged and older men more often than women with debilitating diseases. This patient population is believed to have impaired respiratory host defenses, including persons with diabetes, alcoholism, malignancy, liver disease, chronic obstructive pulmonary diseases, glucocorticoid therapy, kidney failure, and certain occupational exposures (such as papermill workers). Many of these infections are obtained when a person is in the hospital for some other reason (a nosocomial infection).
In addition to pneumonia, Klebsiella can also cause infections in the urinary tract, lower biliary tract, and surgical wound sites. The range of clinical diseases includes pneumonia, thrombophlebitis, urinary tract infection, cholecystitis, diarrhea, upper respiratory tract infection, wound infection, osteomyelitis, meningitis, and bacteremia, and sepsis. For patients with an invasive device in their bodies, contamination of the device becomes a risk; neonatal ward devices, respiratory support equipment, and urinary catheters put patients at increased risk. Also, the use of antibiotics can be a factor that increases the risk of nosocomial infection with Klebsiella bacteria. Sepsis and septic shock can follow entry of the bacteria into the blood.
Research conducted at King's College, London has implicated molecular mimicry between HLA-B27 and two Klebsiella surface molecules as the cause of ankylosing spondylitis.[5]
Klebsiella ranks second to E. coli for urinary tract infections in older people.[6] It is also an opportunistic pathogen for patients with chronic pulmonary disease, enteric pathogenicity, nasal mucosa atrophy, and rhinoscleroma. New antibiotic-resistant strains of K. pneumoniae are appearing.[7]
The most common condition caused by Klebsiella bacteria outside the hospital is pneumonia, typically in the form of bronchopneumonia and also bronchitis. These patients have an increased tendency to develop lung abscesses, cavitation, empyema, and pleural adhesions. It has a death rate around 50%, even with antimicrobial therapy. [8]
It is typically due to aspiration and alcoholism may be a risk factor, though it is also commonly implicated in hospital-acquired urinary tract infections, and COPD (chronic obstructive pulmonary disease) individuals.[9][10] In terms of the pathophysiology of Klebsiella pneumonia the neutrophil myeloperoxidase defense against K. pneumoniae is often seen. Oxidative inactivation of elastase is involved, while LBP helps transfer bacteria cell wall elements to the cells.[11][12]
Individuals with Klebsiella pneumoniae tend to cough up a characteristic sputum, as well as having fever, nausea, tachycardia, and vomiting. Klebsiella pneumoniae tends to affect people with underlying conditions, such as alcoholism.[9]
In terms of the diagnosis of Klebsiella pneumoniae the following can be done to determine if the individual has this infection, with the addition of susceptibility testing to identify drug-resistant organisms:[11][9]
Treatment for Klebsiella pneumoniae is by antibiotics such as aminoglycosides, pipercillin tazobactam, and cephalosporins, the choice depending upon antibiotic susceptibility testing, the person's health condition, medical history and severity of the disease.[10][13]
Klebsiella possesses beta-lactamase giving it resistance to ampicillin, many strains have acquired an extended-spectrum beta-lactamase with additional resistance to carbenicillin, amoxicillin, and ceftazidime. The bacteria remain susceptible to aminoglycosides and some cephalosporins, and varying degrees of inhibition of the beta-lactamase with clavulanic acid have been reported. Infections due to multidrug-resistant gram-negative pathogens in the ICU have invoked the re-emergence of colistin. However, colistin-resistant strains of K. pneumoniae have been reported in ICUs.[11][14][15][16] In 2009, strains of K. pneumoniae with gene called New Delhi metallo-beta-lactamase ( NDM-1) that even gives resistance against intravenous antibiotic carbapenem, were discovered in India and Pakistan. Klebsiella cases in Taiwan have shown abnormal toxicity, causing liver abscesses in people with diabetes mellitus (DM); treatment consists of third generation cephalosporins.
Hypervirulent (hvKp) is a rather recent K pneumoniae variant that is significantly more virulent than classical K. pneumoniae (cKp). While cKp is an opportunistic pathogen responsible for nosocomial infections that usually affect immunocompromised patients, hvKp is clinically more concerning since it also causes disease in healthy individuals and can infect virtually every site of the body. The genetic traits that lead to this pathotype are included in a large virulence plasmid and potentially on additional conjugative elements.[17]
These newly identified strains were described to overproduce capsule components and siderophores for iron acquisition, among other factors.[18] Although initial studies showed that hvKp is rather susceptible to antibiotic treatment, it has been recently shown that such strains can acquire resistance plasmids and become multiresistant to a variety of antibiotics.[18][19][20]
It is originated from Asia, having a high mortality rate among the population. It often spreads to central nervous system and eye causing endophthalmitis, nonhepatic abscesses, pneumonia, necrotizing fasciitis, and meningitis. One visual trait of these strains is hypermucoviscous phenotype and a string test can be used to help the diagnosis.[21] Further examinations and treatments are made on a case-by-case basis, as there are currently no international guidelines.[22]
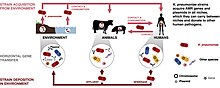
To get a K. pneumoniae infection, a person must be exposed to the bacteria. In other words, K. pneumoniae must enter the respiratory tract to cause pneumonia, or the blood to cause a bloodstream infection. In healthcare settings, K. pneumoniae bacteria can be spread through person-to-person contact (for example, contaminated hands of healthcare personnel, or other people via patient to patient) or, less commonly, by contamination of the environment; the role of transmission directly from the environment to patients is controversial and requires further investigation.[23] However, the bacteria are not spread through the air. Patients in healthcare settings also may be exposed to K. pneumoniae when they are on ventilators, or have intravenous catheters or wounds. These medical tools and conditions may allow K. pneumoniae to enter the body and cause infection.[24]
Klebsiella organisms are often resistant to multiple antibiotics. Current evidence implicates plasmids as the primary source of the resistance genes.[25] Klebsiella species with the ability to produce extended-spectrum beta-lactamases (ESBL) are resistant to virtually all beta-lactam antibiotics, except carbapenems. Other frequent resistance targets include aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol, and trimethoprim/sulfamethoxazole.[26]

Infection with carbapenem-resistant Enterobacteriaceae (CRE) or carbapenemase-producing Enterobacteriaceae is emerging as an important challenge in health-care settings.[28][29] One of many CREs is carbapenem-resistant Klebsiella pneumoniae (CRKP). Over the past 10 years, a progressive increase in CRKP has been seen worldwide; however, this new emerging nosocomial pathogen is probably best known for an outbreak in Israel that began around 2006 within the healthcare system there.[30] In the US, it was first described in North Carolina in 1996;[31] since then CRKP has been identified in 41 states;[32] and is routinely detected in certain hospitals in New York and New Jersey. It is now the most common CRE species encountered within the United States.
CRKP is resistant to almost all available antimicrobial agents, and infections with CRKP have caused high rates of morbidity and mortality, in particular among persons with prolonged hospitalization and those critically ill and exposed to invasive devices (e.g., ventilators or central venous catheters). The concern is that carbapenem is often used as a drug of last resort when battling resistant bacterial strains. New slight mutations could result in infections for which healthcare professionals can do very little, if anything, to treat patients with resistant organisms.
A number of mechanisms cause carbapenem resistance in the Enterobacteriaceae. These include hyperproduction of ampC beta-lactamase with an outer membrane porin mutation, CTX-M extended-spectrum beta-lactamase with a porin mutation or drug efflux, and carbapenemase production. The most important mechanism of resistance by CRKP is the production of a carbapenemase enzyme, blakpc. The gene that encodes the blakpc enzyme is carried on a mobile piece of genetic material (a transposon; the specific transposon involved is called Tn4401), which increases the risk for dissemination. CRE can be difficult to detect because some strains that harbor blakpc have minimum inhibitory concentrations that are elevated, but still within the susceptible range for carbapenems. Because these strains are susceptible to carbapenems, they are not identified as potential clinical or infection control risks using standard susceptibility testing guidelines. Patients with unrecognized CRKP colonization have been reservoirs for transmission during nosocomial outbreaks.[33]
The extent and prevalence of CRKP within the environment is currently unknown. The mortality rate is also unknown, but has been observed to be as high as 44%.[34] The Centers for Disease Control and Prevention released guidance for aggressive infection control to combat CRKP:
Israel 2007-2008. A nationwide outbreak of CRE in Israel peaked in March, 2007 at 55.5 cases per 100,000 patient days and necessitated a nationwide treatment plan. The intervention entailed physical separation of all CRE carriers and appointment of a task force to oversee efficacy of isolation by closely monitoring hospitals and intervening when necessary. After the treatment plan (measured in May, 2008), the number of cases per 100,000 patient days decreased to 11.7. The plan was effective because of strict hospital compliance, wherein each was required to keep detailed documentation of all CRE carriers. In fact, for each increase in compliance by 10%, incidence of cases per 100,000 patient days decreased by 0.6. Therefore, containment on a nationwide scale requires nationwide intervention.[36]
Nevada 2016. In mid-August 2016, a resident of Washoe County was hospitalized in Reno due to a CRE (specifically Klebsiella pneumoniae) infection. In early September of the same year, she developed septic shock and died. On testing by CDC an isolate from the patient was found to be resistant to all 26 antibiotics available in the US, including drug of last resort colistin.[37] It is believed she may have picked up the microbe while hospitalized in India for two years due to a broken right femur and subsequent femur and hip infections.[38][39][40]
Klebsiella pneumoniae carries a large number of anti-microbial resistance genes (AMR genes). These genes are transferred via plasmids from and to other human pathogens. One human pathogen that commonly acquires AMR genes from Klebsiella pneumoniae is Salmonella. This could help with treatment of salmonella infections due to having knowledge of possible antibiotic resistance data.
The majority of AMR genes in Klebsiella pneumoniae are plasmid-borne. An example of a niche would be soil, often considered a hotspot for gene transfer.
The table shows the number of AMR genes and plasmids (per strain or subspecies) compared to other common bacteria species.[41]
To prevent spreading Klebsiella infections between patients, healthcare personnel must follow specific infection-control precautions,[24] which may include strict adherence to hand hygiene (preferably using an alcohol based hand rub (60-90%) or soap and water if hands are visibly soiled. Alcohol based hand rubs are effective against these Gram-negative bacilli)[42] and wearing gowns and gloves when they enter rooms where patients with Klebsiella–related illnesses are housed. Healthcare facilities also must follow strict cleaning procedures to prevent the spread of Klebsiella.[24]
To prevent the spread of infections, patients also should clean their hands very often, including:
K. pneumoniae can be treated with antibiotics if the infections are not drug-resistant. Infections by K. pneumoniae can be difficult to treat because fewer antibiotics are effective against them. In such cases, a microbiology laboratory must run tests to determine which antibiotics will treat the infection.[24] More specific treatments of Klebsiella pneumonia are given in its section above. For urinary tract infections with multidrug-resistant Klebsiella species, a combination therapy with amikacin and meropenem has been suggested.[43]
Multiple drug-resistant K. pneumoniae strains have been killed in vivo by intraperitoneal, intravenous, or intranasal administration of phages in laboratory tests.[44] Resistance to phages is not likely to be as troublesome as to antibiotics as new infectious phages are likely to be available in environmental reservoirs. Phage therapy can be used in conjunction with antibiotics, to supplement their activity instead of replacing it altogether.[45]
New data sources outlining the global burden of K. pneumoniae and drug-resistant forms are expected to build momentum into prophylactic vaccine development.[46] The recent 2022 IHME study showed that in 2019 K. pneumoniae was responsible for 790,000 deaths [571,000 - 1,060,000] in all age groups across 11 infectious syndromes. Importantly, in Sub-saharan Africa K. pneumoniae was responsible for 124,000 [89,000-167,000] neonatal deaths due to bloodstream infections. Based on these and other data, a newly developed prophylactic vaccine would ideally be designed to prevent invasive K. pneumoniae disease in both vulnerable persons but also as a maternal vaccine to prevent neonatal sepsis and global demand assessments have been published.[47] As of June 2023, one single clinical development program for a K. pneumoniae vaccine [Kleb4V/GSK4429016A] was in a Phase 1/2 study in healthy adults aged 18-70 yrs (n=166) [Clinical trials identifier: NCT04959344]. The vaccine is an O-antigen based conjugate where the specific O-antigens in the vaccine remain undisclosed [Michael Kowarik, LimmaTech Biologics, World Vaccine Congress EU, 2022] although only a limited number of O-serotypes can account for a high proportion of clinical isolates. [48]
Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated, lactose-fermenting, facultative anaerobic, rod-shaped bacterium. It appears as a mucoid lactose fermenter on MacConkey agar.
Although found in the normal flora of the mouth, skin, and intestines, it can cause destructive changes to human and animal lungs if aspirated, specifically to the alveoli resulting in bloody, brownish or yellow colored jelly-like sputum. In the clinical setting, it is the most significant member of the genus Klebsiella of the Enterobacteriaceae. K. oxytoca and K. rhinoscleromatis have also been demonstrated in human clinical specimens. In recent years, Klebsiella species have become important pathogens in nosocomial infections.
It naturally occurs in the soil, and about 30% of strains can fix nitrogen in anaerobic conditions. As a free-living diazotroph, its nitrogen-fixation system has been much-studied, and is of agricultural interest, as K. pneumoniae has been demonstrated to increase crop yields in agricultural conditions.
It is closely related to K. oxytoca from which it is distinguished by being indole-negative and by its ability to grow on melezitose but not 3-hydroxybutyrate.