-
Daisuke Uyeno, Kazuya Nagasawa
Zookeys
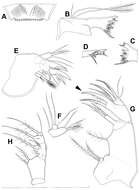
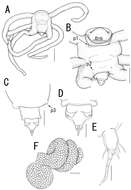
Figure 6.Splanchnotrophus helianthus sp. n., female, holotype NSMT–Cr 22244. A antennule, anterior B oral area C mandible, posterior D maxilla E leg 1 F leg 2 G leg 3. Scale bars = 20 μm in A, E, F; 30 μm in B; 10 μm in C, D, G.
-
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
Figure 16.Diacyclops suoensis Ito, 1954, female: A labrum, anterior view B mandibula, anterior view C cutting edge of mandibula, posterior view D cutting edge of mandibula, dorsal view E maxillula, posterior view F maxillular palp, posterior view G maxilla, anterior view H maxilliped, anterior view. Arrows pointing most prominent specific features. Scale bar 100 μm.
-
Daisuke Uyeno, Kazuya Nagasawa
Zookeys
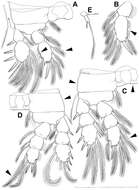
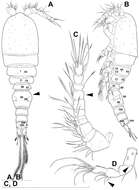
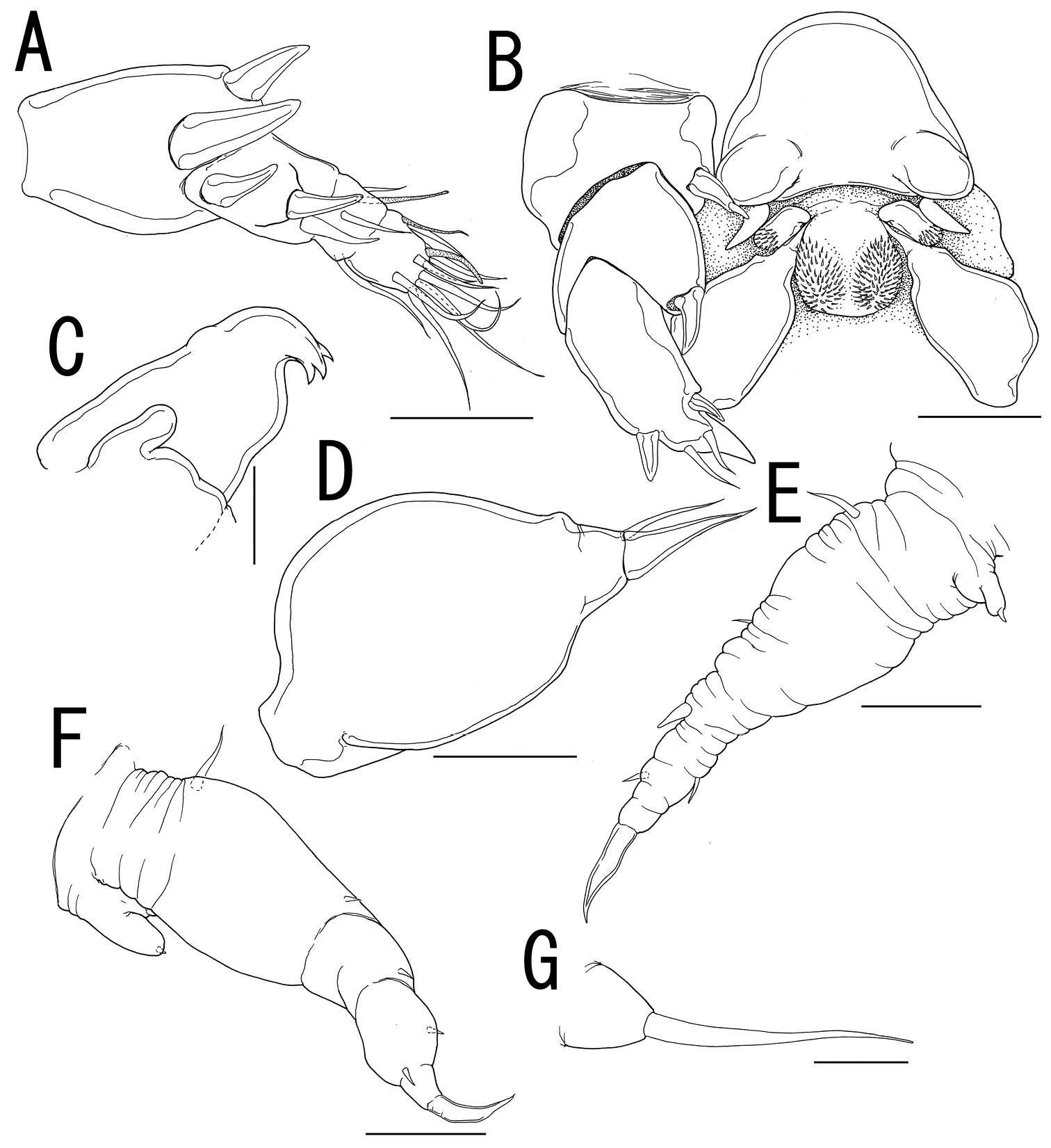
Figure 7.Splanchnotrophus helianthus sp. n., male, allotype NSMT–Cr 22245. A habitus, dorsal B habitus, ventral C habitus, lateral D free thoracic somites and abdomen, ventral E caudal ramus, ventral F oral area G leg 1 H leg 2. Scale bars = 100 μm in A, B, C; 50 μm in D; 20 μm in E.
-
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
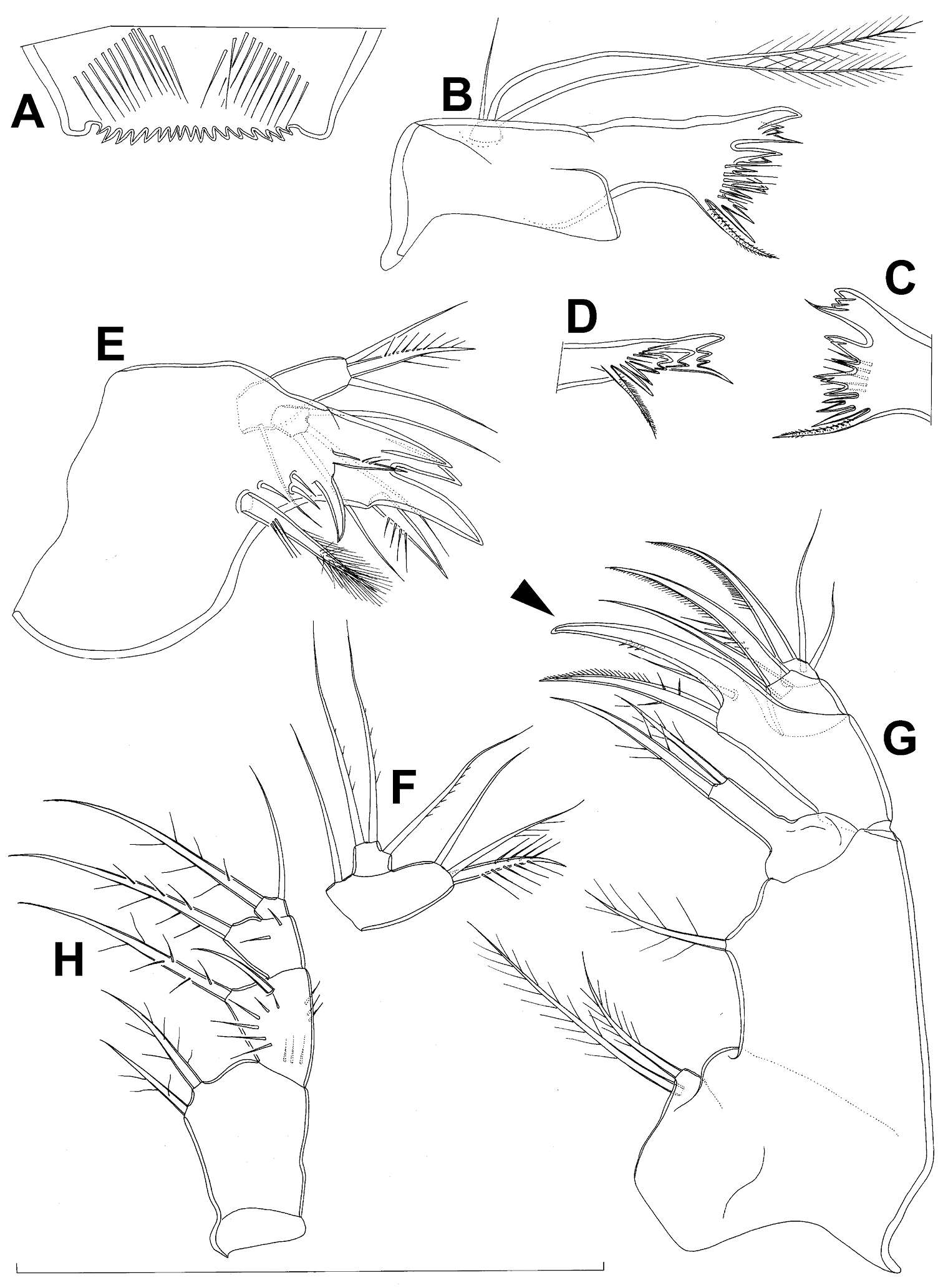
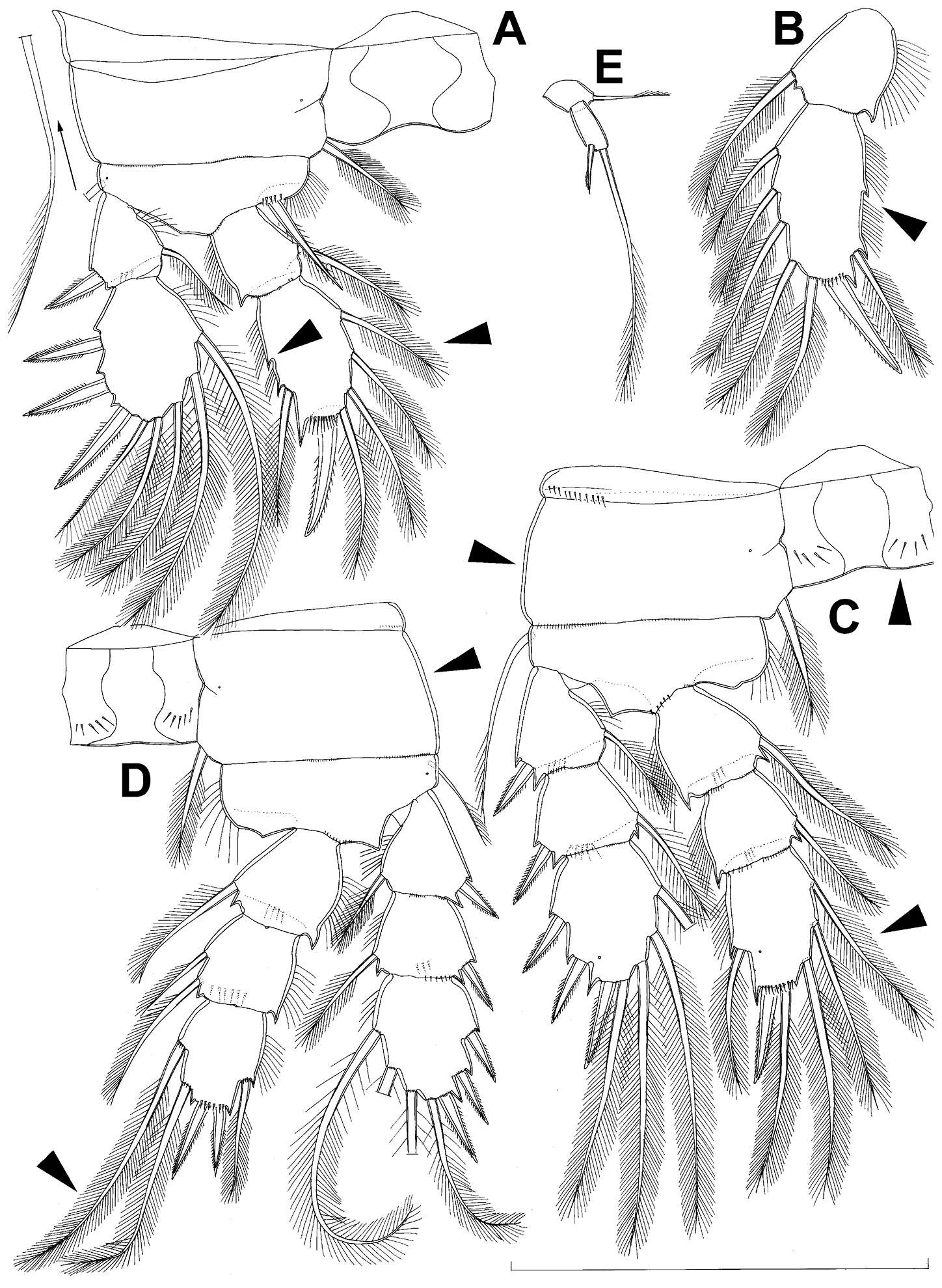
Figure 17.Diacyclops suoensis Ito, 1954, female: A first swimming leg, anterior view B endopod of second swimming leg, anterior view C third swimming leg, anterior view D fourth swimming leg, anterior view E fifth leg, anterior view. Arrows pointing most prominent specific features. Scale bar 100 μm.
-
Daisuke Uyeno, Kazuya Nagasawa
Zookeys
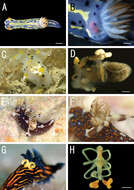
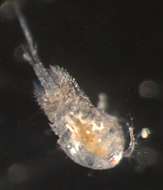
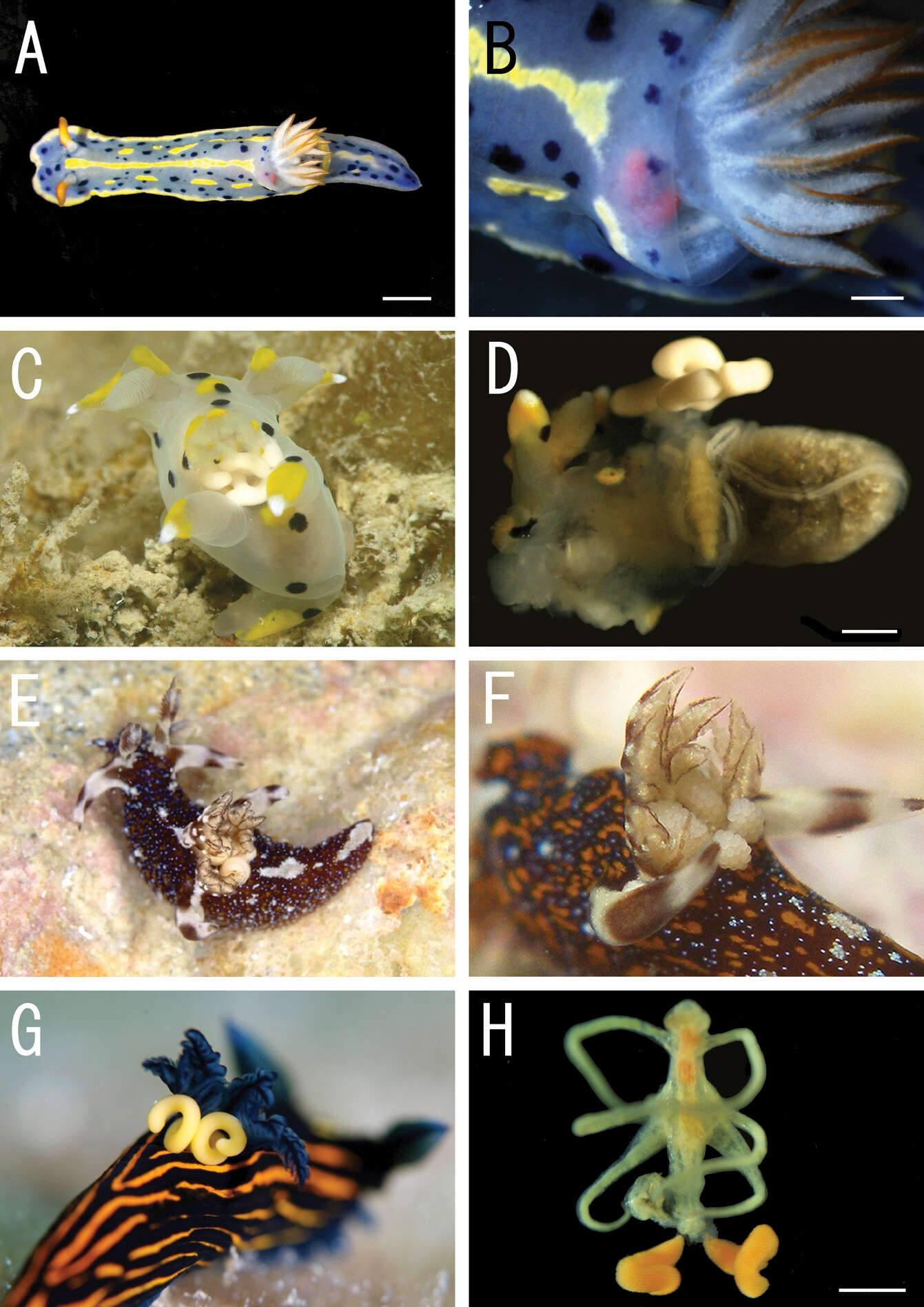
Figure 1.Live coloration of the host nudibranchs and the splanchnotrophids. A Hypselodoris festiva infected by an ovigerous specimenof Certosomicola japonica sp. n. B an egg sac of Ceratosomicola japonica sp. n. and the gill circle of Hypselodoris festiva with the mantle malformed into an elongate tube C Thecacera pennigera infected by an ovigerous specimen of Splanchnotrophus helianthus sp. n. D Trapania pennigera with the mantle removed to show a female specimen of Splanchnotrophus helianthus on the visceral sac E Trapania miltabrancha infected by an ovigerous specimen of Splanchnotrophus imagawai sp. n (photo by K. Imagawa) F gill circle of Trapania miltabrancha with egg sacs of Splanchnotrophus imagawai sp. n. (photo by K. Imagawa) G Roboastra luteolineata infected by an ovigerous specimen of Majimun shirakawai gen. et sp. n. (photo by N. Shirakawa) H female Majimun shirakawai gen. et sp. n. with dwarf male attached to the posterior part of the body. Scale bars = 5 mm in A; 1 mm in B, D, H.
-
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
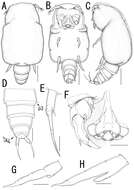
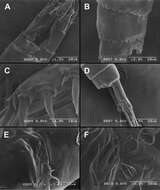
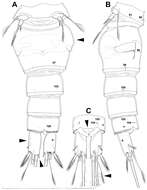
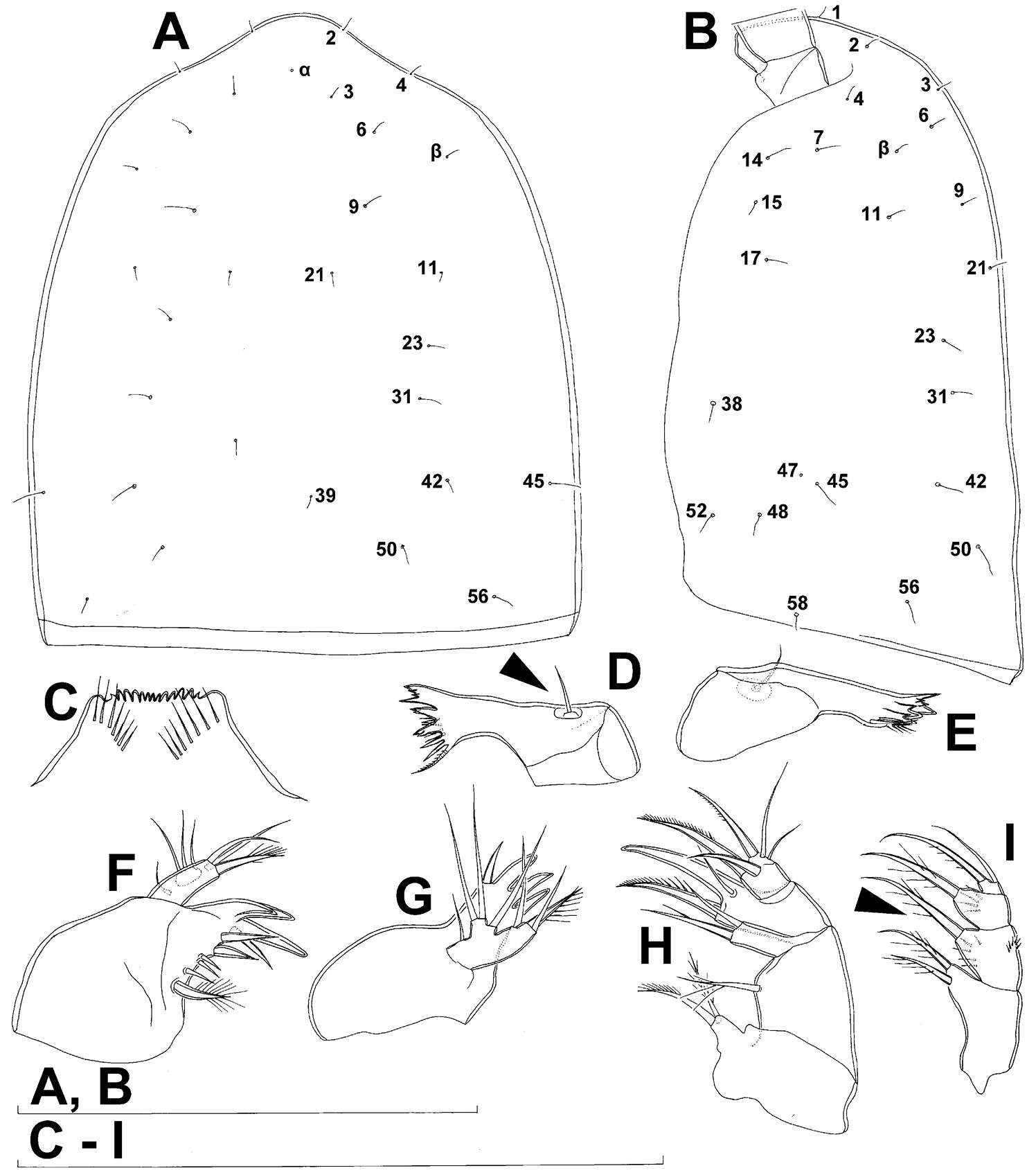
Figure 26.Scanning electron micrographs, A–C Diacyclops ishidai sp. n. D–E Diacyclops parasuoensis sp. n. F Diacyclops suoensis Ito, 1954: A anal somite and caudal rami, dorsal view, paratype female 1 B preanal and anal somites, lateral view, paratype female 2 C last two exopodal segments of second swimming legs, lateral view, paratype female 2 D anal somite and caudal rami, lateral view, paratype female E sixth leg, lateral view, paratype female F labrum and maxillulae, ventral view. Scale bars 20 μm (A, B, D, F) and 10 μm (C, E).
-
Daisuke Uyeno, Kazuya Nagasawa
Zookeys
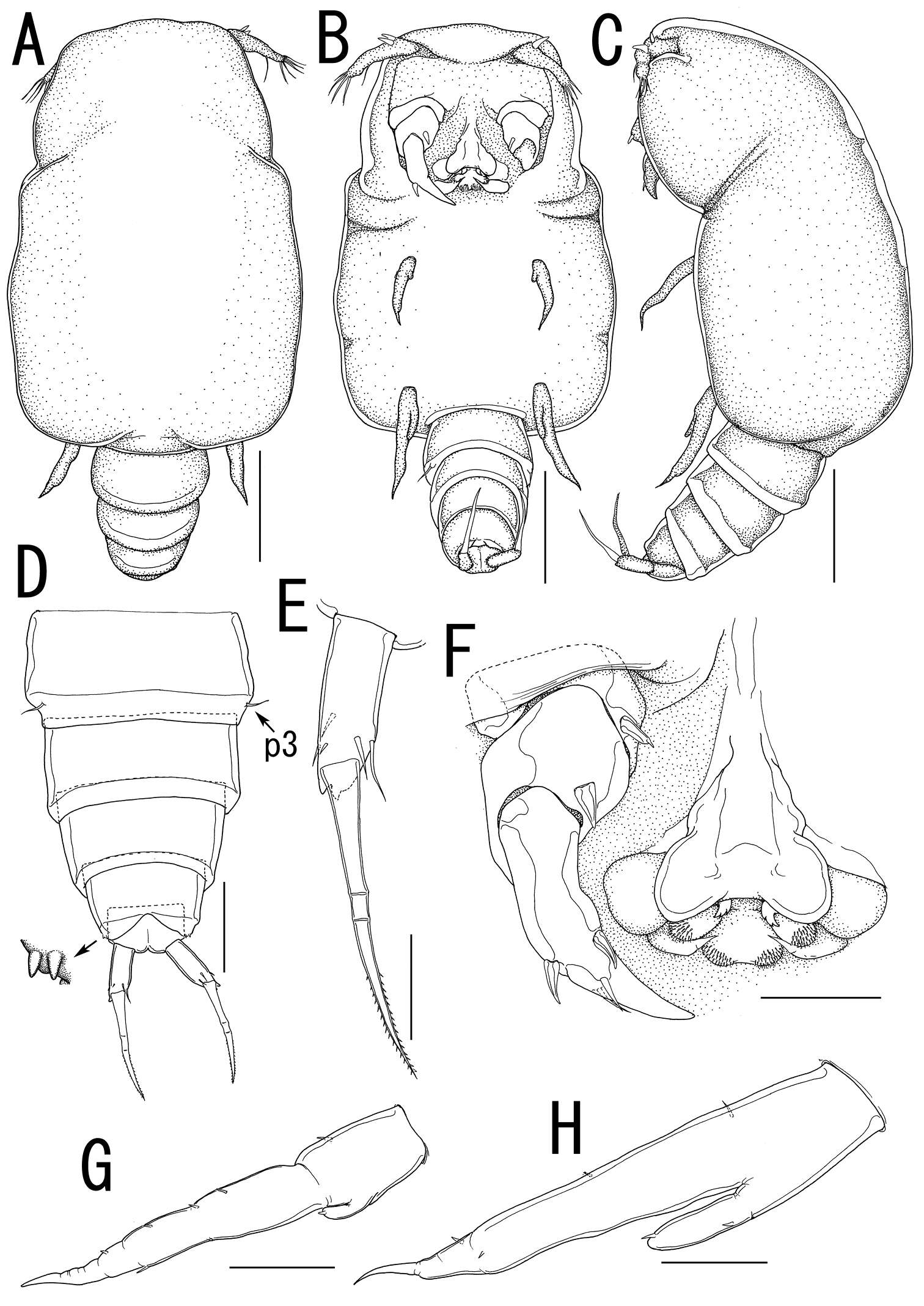
Figure 8.Splanchnotrophus imagawai sp. n., female, holotype NSMT–Cr 22249. A habitus, dorsal B habitus, ventral, p1 = leg 1, p2 = leg 2 C posterior portion of body, ventral, p3 = leg 3 D fourth pedigerous somite and genito-abdomen, ventral E caudal ramus, ventral F egg sac. Scale bars = 1 mm in A; 500 μm in B, F; 200 μm in C; 100 μm in D; 20 μm in E.
-
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
Figure 18.Diacyclops hanguk sp. n., holotype female: A habitus, dorsal view B habitus, lateral view C antennula, dorsal view D antenna, ventral view. Arabic numerals indicating sensilla and pores presumably homologous to those in Diacyclops ishidai sp. n. Roman numerals indicating pores homologous to those in Diacyclops parasuoensis sp. n. Arrows pointing most prominent specific features. Scale bars 100 μm.
-
Daisuke Uyeno, Kazuya Nagasawa
Zookeys
Figure 9.Splanchnotrophus imagawai sp. n., female, holotype NSMT–Cr 22249. A anterior portion of cephalosome B antennule, ventral C oral area D mandible, posterior E maxilla F leg 1 G leg 2 H leg 3. Scale bars = 100 μm in A; 50 μm in B, C; 10 μm in D, H; 20 μm in E, F, G.
-
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
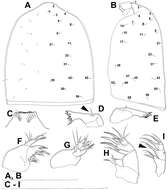
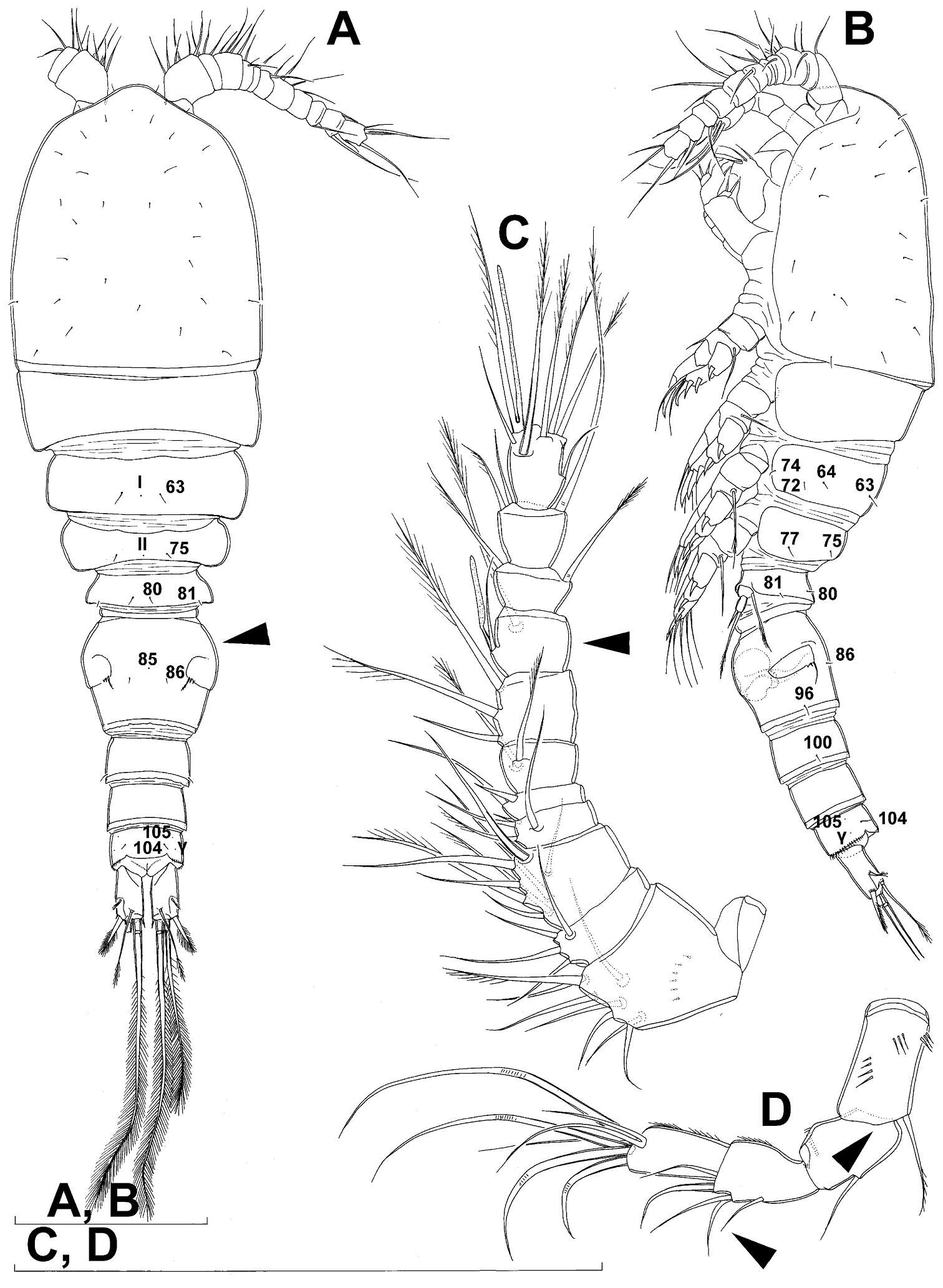
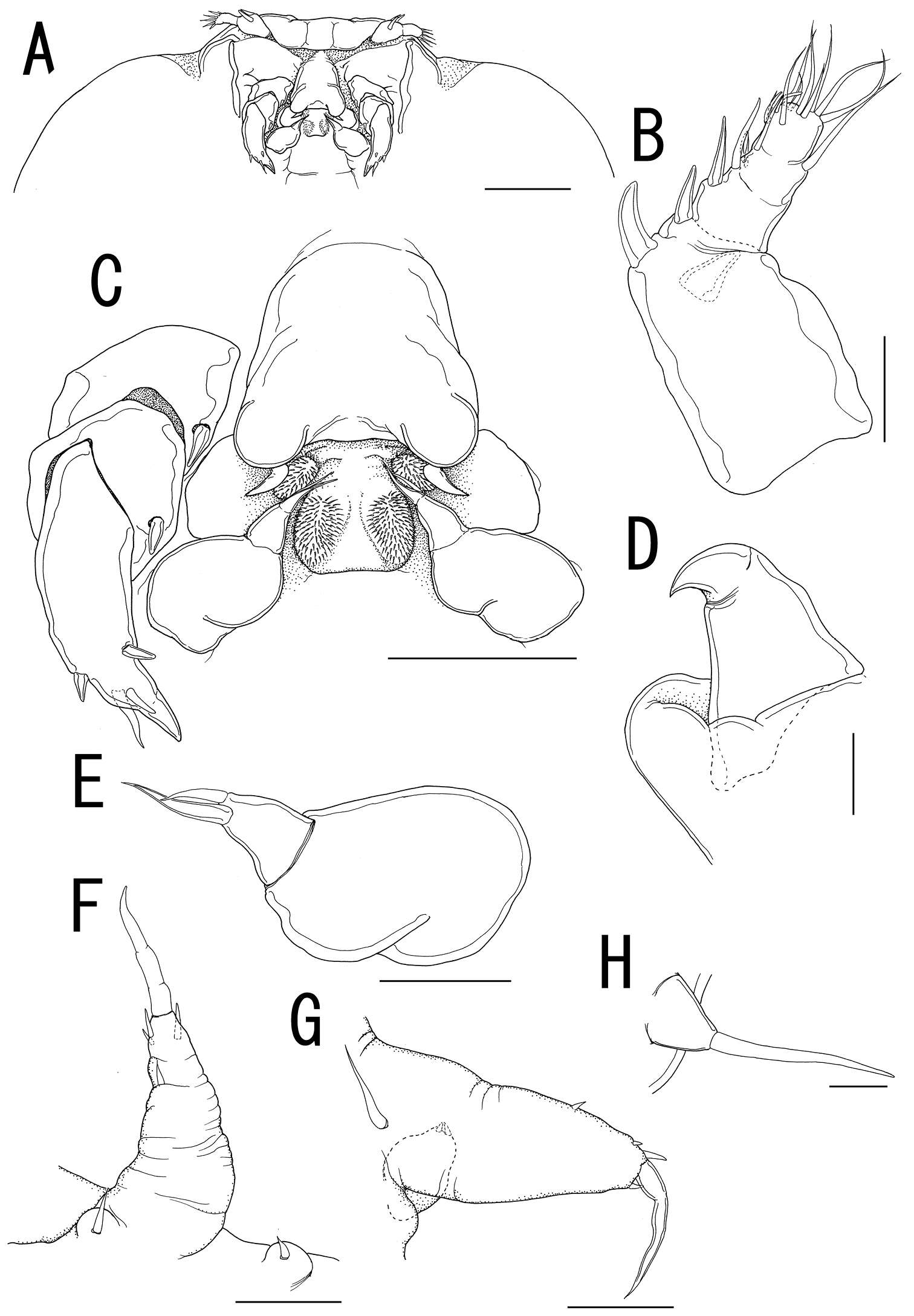
Figure 19.Diacyclops hanguk sp. n., holotype female: A cephalothorax, dorsal view B cephalothoracic shield, lateral view C labrum, anterior view D mandibula, posterior view E mandibula, antero-ventral view F maxillula, posterior view G maxillula, anterior view H maxilla, posterior view I maxilliped, posterior view. Arabic numerals indicating sensilla and pores presumably homologous to those in Diacyclops ishidai sp. n. Greek letters indicating unique pores and sensilla. Arrows pointing most prominent specific features. Scale bars 100 μm.
-
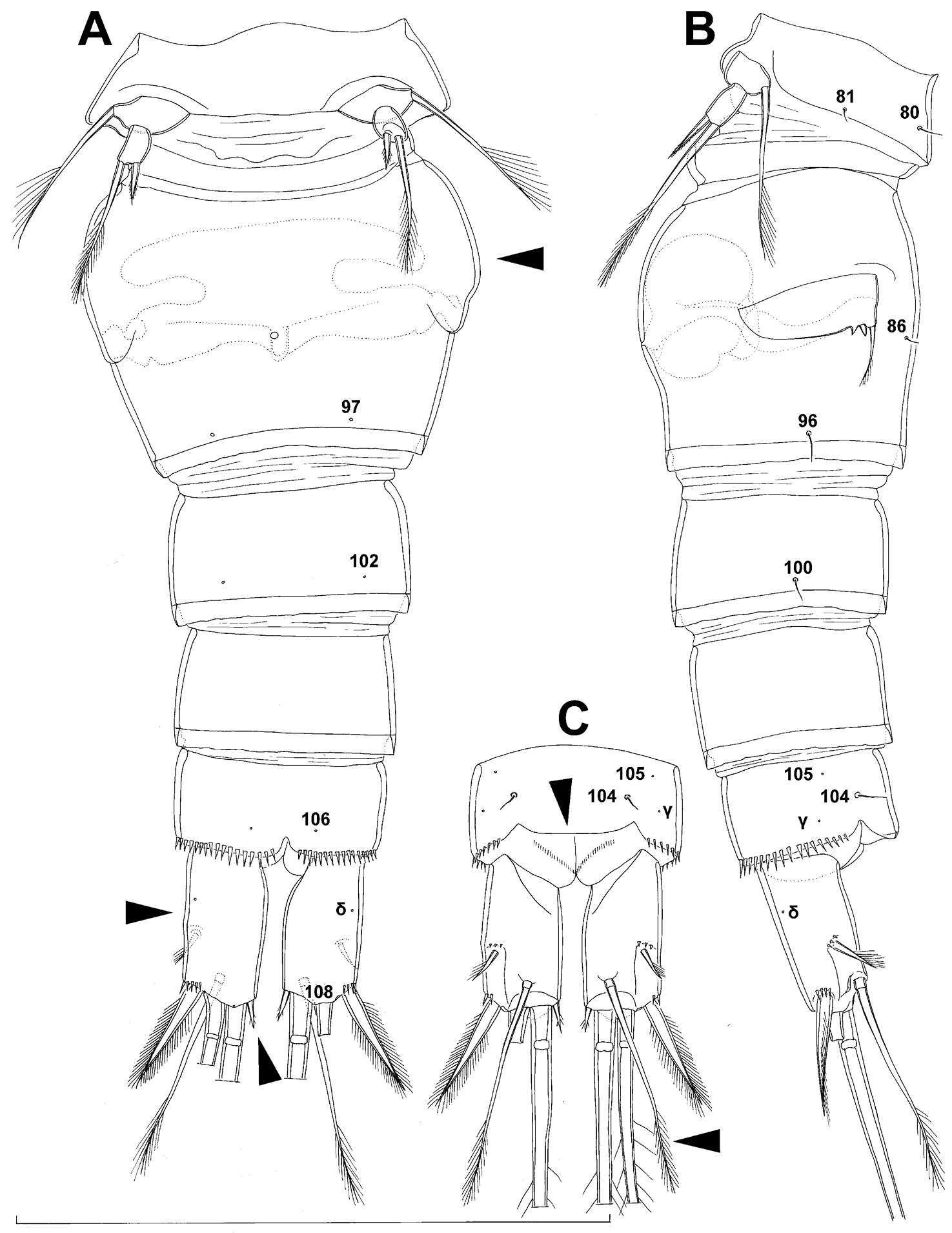
Tomislav Karanovic, Mark J. Grygier, Wonchoel Lee
Zookeys
Figure 20.Diacyclops hanguk sp. n., holotype female: A urosome, ventral view B usorome, lateral view C anal somite and caudal rami, dorsal view. Arabic numerals indicating sensilla and pores presumably homologous to those in Diacyclops ishidai sp. n. Greek letters indicating unique pores and sensilla. Arrows pointing most prominent specific features. Scale bar 100 μm.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.