-
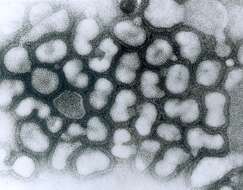
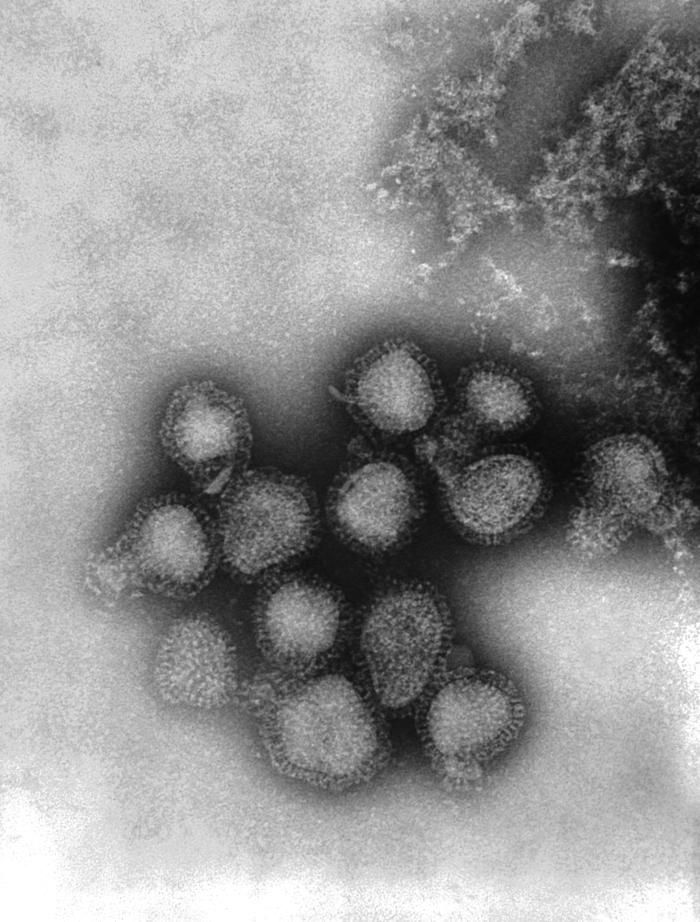
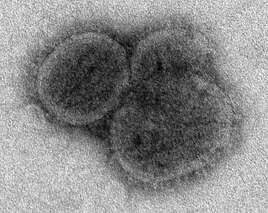
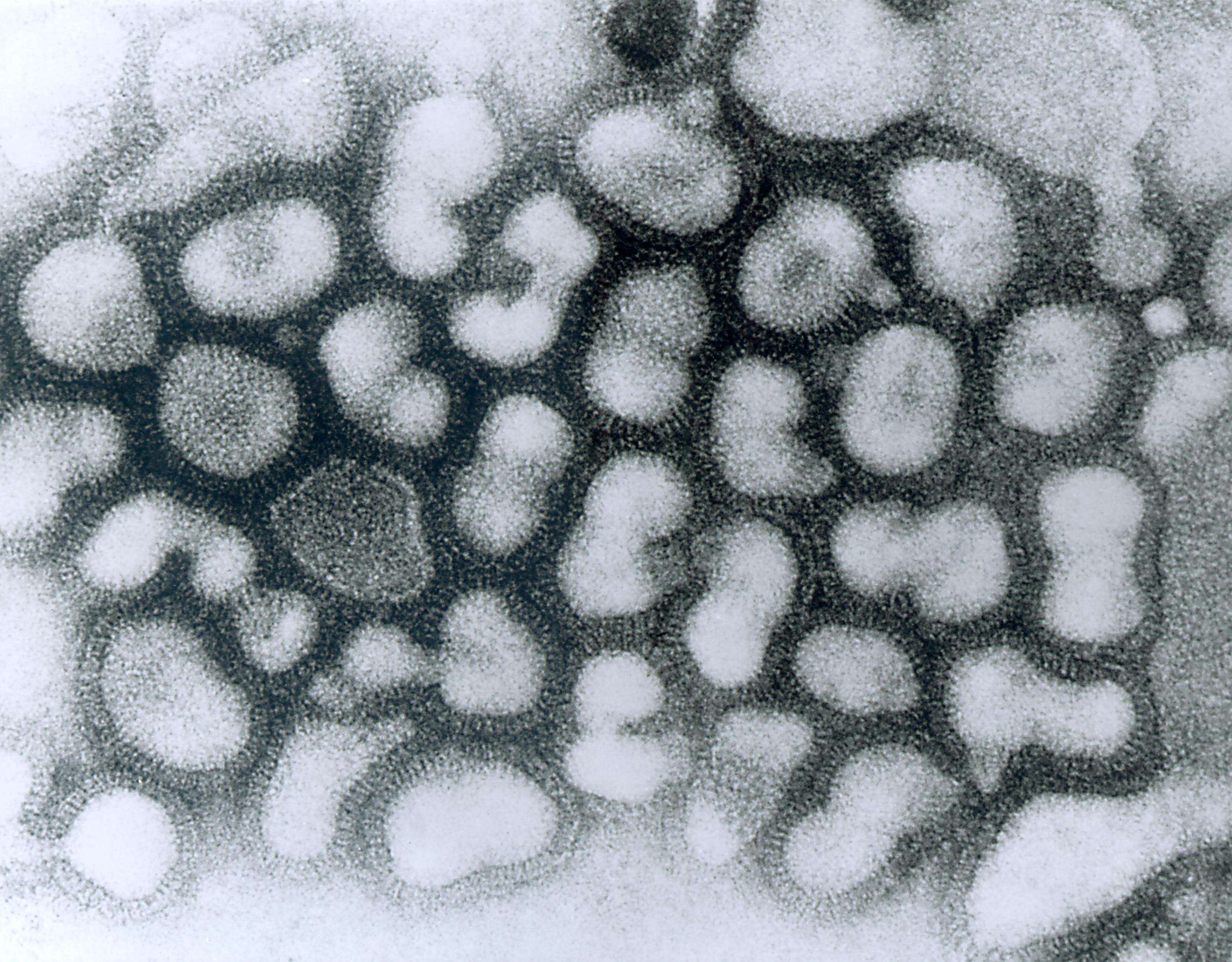
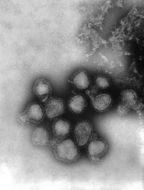
This negatively-stained transmission electron micrograph (TEM) revealed the presence of a number of influenza virus virions. This virus is a Orthomyxoviridae virus family member.Created: 1975
-
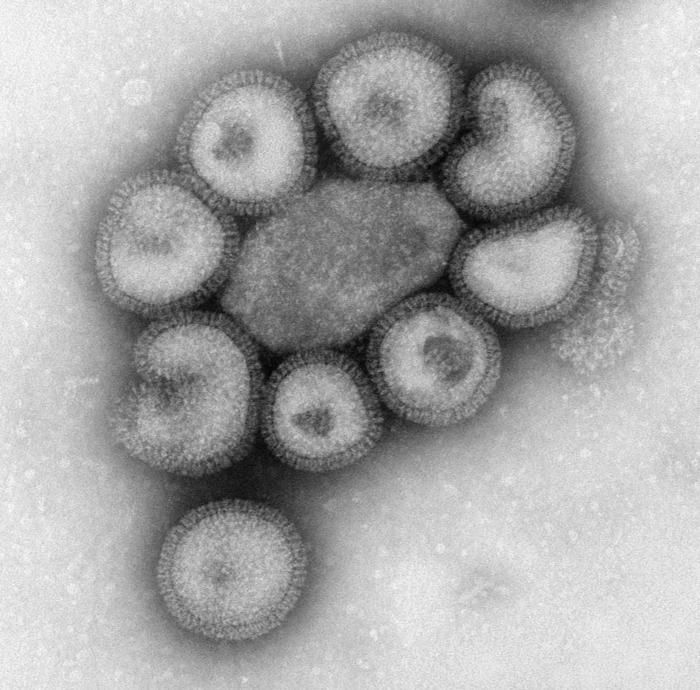
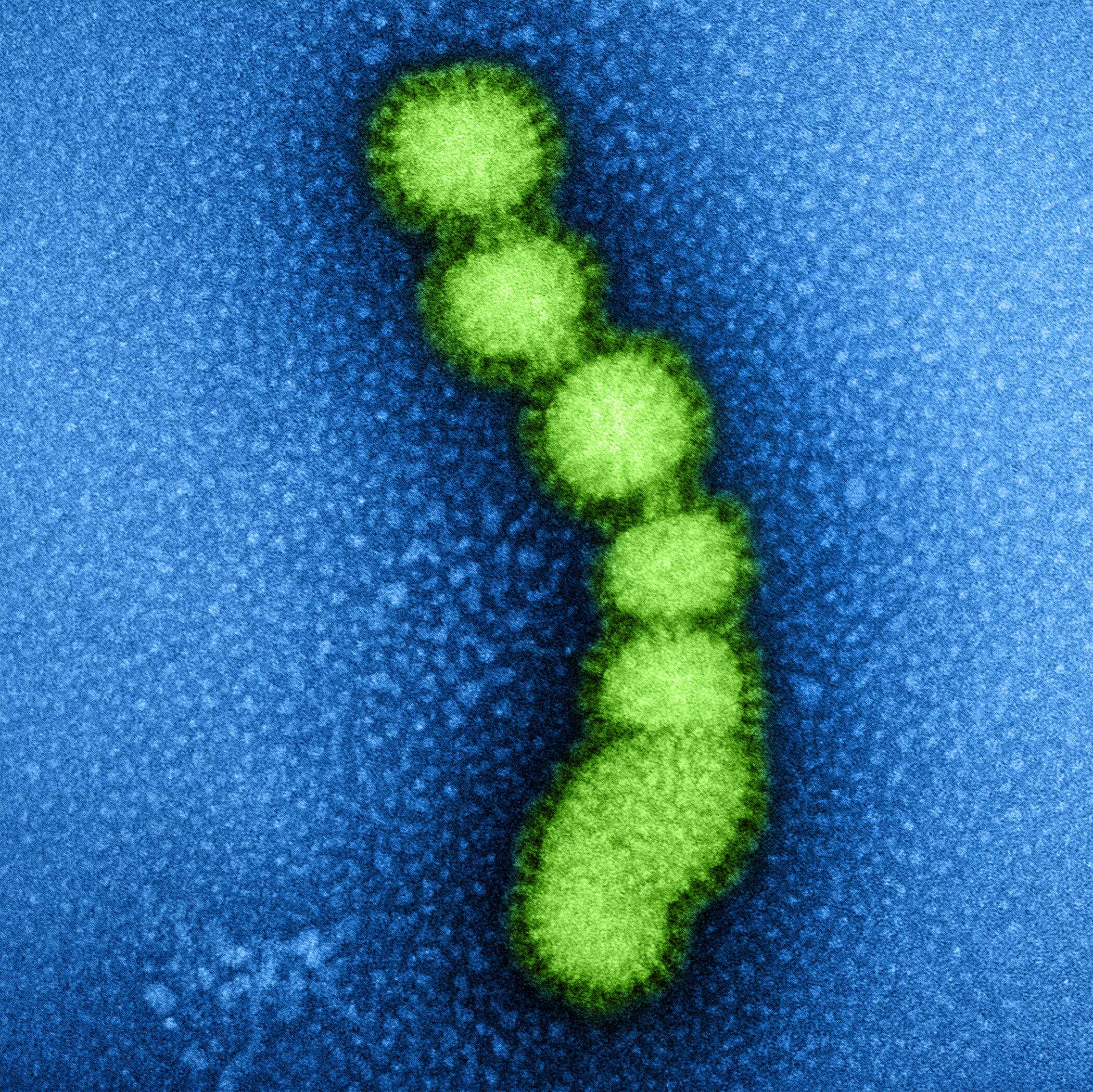
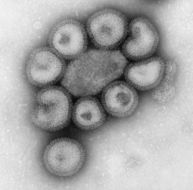
This negatively-stained transmission electron micrograph (TEM) revealed the presence of a number of Hong Kong flu virus virions, the H3N2 subtype of the influenza A virus. This virus is a Orthomyxoviridae virus family member, and was responsible for the flu pandemic of 1968-1969, which infected an estimated 50,000,000 people in the United States, killing 33,000. Note the proteinaceous coat, or capsid, surrounding each virion, and the hemagglutinin-neuraminidase spikes, which differ in terms of their molecular make-up from strain to strain.Created: 1975
-
This negatively-stained transmission electron micrograph (TEM) revealed the presence of a number of Hong Kong flu virus virions, the H3N2 subtype of the influenza A virus. This virus is a Orthomyxoviridae virus family member, and was responsible for the flu pandemic of 1968-1969, which infected an estimated 50,000,000 people in the United States, killing 33,000. Note the proteinaceous coat, or capsid, surroundind each virion, and the hemagglutinin-neuraminidase spikes, which differ in terms of their molecular make-up from strain to strain.Created: 1975
-
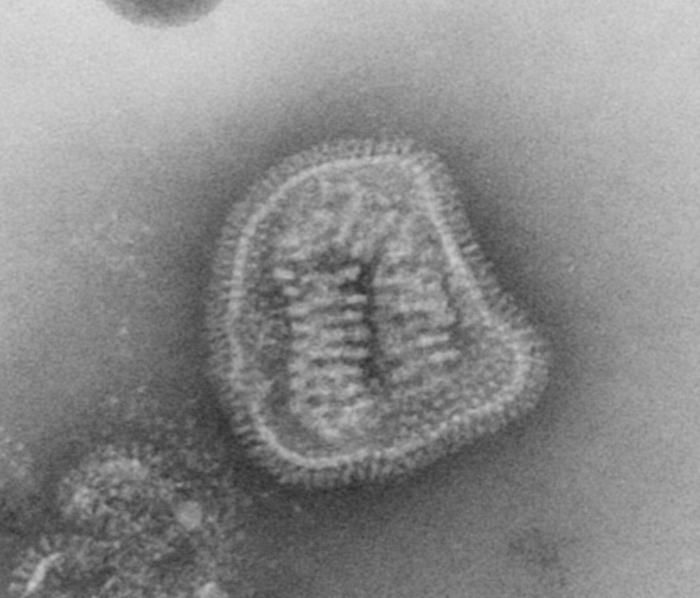
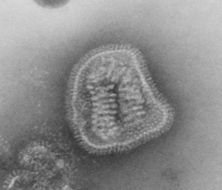
This negative-stained transmission electron micrograph (TEM) depicts the ultrastructural details of a number of influenza virus particles, or virions. A member of the taxonomic family Orthomyxoviridae, the influenza virus is a single-stranded RNA organismThe flu is a contagious respiratory illness caused by influenza viruses. It can cause mild to severe illness, and at times can lead to death. The best way to prevent this illness is by getting a flu vaccination each fall.Every year in the United States, on average:- 5% to 20% of the population gets the flu- more than 200,000 people are hospitalized from flu complications, and- about 36,000 people die from flu. Some people, such as older people, young children, and people with certain health conditions, are at high risk for serious flu complications. See PHIL 10072 for a colorized version of this image.Created: 1973
-
This negative-stained transmission electron micrograph (TEM) depicts the ultrastructural details of an influenza virus particle, or virion. A member of the taxonomic family Orthomyxoviridae, the influenza virus is a single-stranded RNA organismThe flu is a contagious respiratory illness caused by influenza viruses. It can cause mild to severe illness, and at times can lead to death. The best way to prevent this illness is by getting a flu vaccination each fall.Every year in the United States, on average:- 5% to 20% of the population gets the flu- more than 200,000 people are hospitalized from flu complications, and- about 36,000 people die from flu. Some people, such as older people, young children, and people with certain health conditions, are at high risk for serious flu complications. See PHIL 10073 for a colorized version of this image.Created: 1981
-
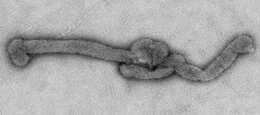
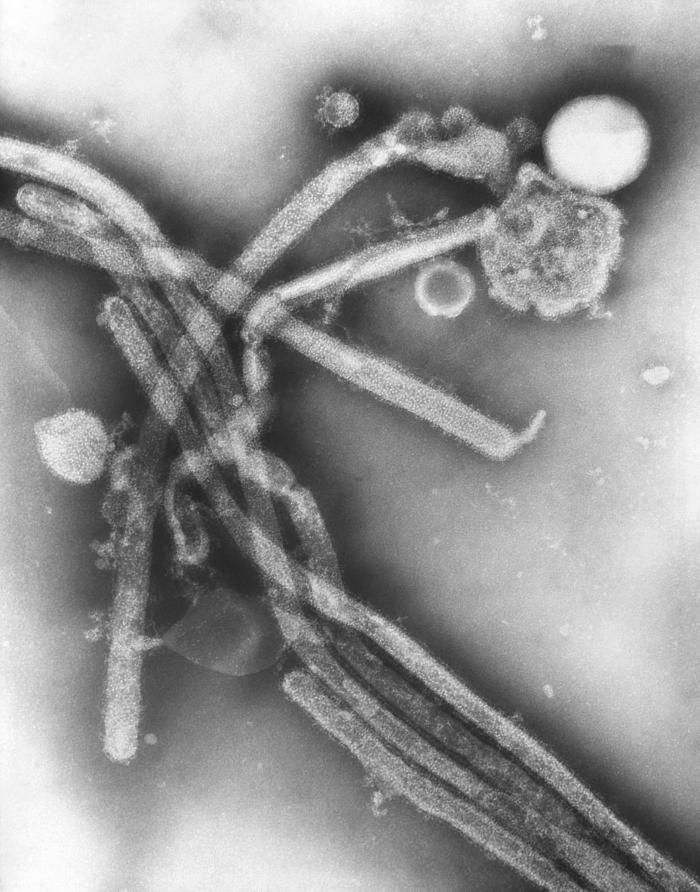
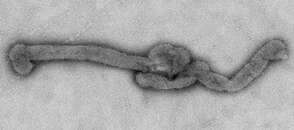
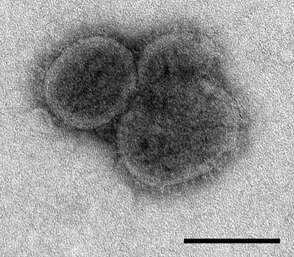
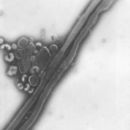
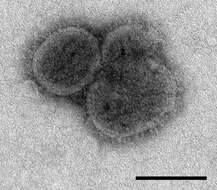
Description: English: Electron microscopic images of novel Thogotovirus isolate, Bourbon virus. Filamentous virus particle with distinct surface projection are visible in culture supernatant that was fixed in 2.5% paraformaldehyde. Cropped from Figure 2a from Kosoy OI, Lambert AJ, Hawkinson DJ, et al. (2015) Novel Thogotovirus species associated with febrile illness and death, United States, 2014. Emerging Infectious Diseases doi:10.3201/eid2105.150150. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
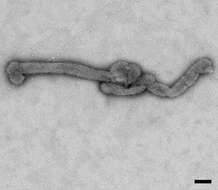
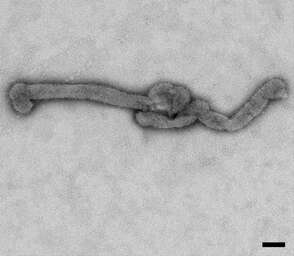
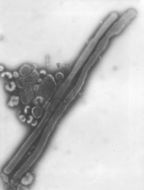
Description: English: Electron microscopic images of novel Thogotovirus isolate, Bourbon virus. Filamentous virus particle with distinct surface projection are visible in culture supernatant that was fixed in 2.5% paraformaldehyde. Scale bar indicates 100 nm. Figure 2a from Kosoy OI, Lambert AJ, Hawkinson DJ, et al. (2015) Novel Thogotovirus species associated with febrile illness and death, United States, 2014. Emerging Infectious Diseases doi:10.3201/eid2105.150150. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
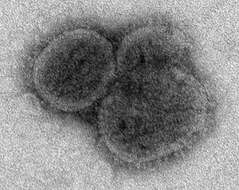
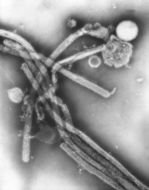
Description: English: Electron microscopic images of novel Thogotovirus isolate, Bourbon virus. Spherical virus particle with distinct surface projection are visible in culture supernatant that was fixed in 2.5% paraformaldehyde. Cropped from Figure 2b from Kosoy OI, Lambert AJ, Hawkinson DJ, et al. (2015) Novel Thogotovirus species associated with febrile illness and death, United States, 2014. Emerging Infectious Diseases doi:10.3201/eid2105.150150. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
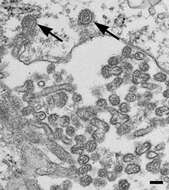
Description: English: Electron microscopic image of novel Thogotovirus isolate, Bourbon virus. Thin-section specimen fixed in 2.5% glutaraldehyde shows numerous extracellular virions with slices through strands of viral nucleocapsids. Arrows indicate virus particles that have been endocytosed. Scale bar indicates 100 nm. Cropped from Figure 2d from Kosoy OI, Lambert AJ, Hawkinson DJ, et al. (2015) Novel Thogotovirus species associated with febrile illness and death, United States, 2014. Emerging Infectious Diseases doi:10.3201/eid2105.150150. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
-
Description: English: Electron microscopic images of novel Thogotovirus isolate, Bourbon virus. Spherical virus particles with distinct surface projection are visible in culture supernatant that was fixed in 2.5% paraformaldehyde. Scale bar indicates 100 nm. Figure 2b from Kosoy OI, Lambert AJ, Hawkinson DJ, et al. (2015) Novel Thogotovirus species associated with febrile illness and death, United States, 2014. Emerging Infectious Diseases doi:10.3201/eid2105.150150. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
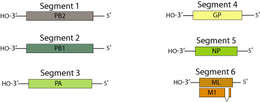
Summary.mw-parser-output table.commons-file-information-table,.mw-parser-output.fileinfotpl-type-information{border:1px solid #a2a9b1;background-color:#f8f9fa;padding:5px;font-size:95%;border-spacing:2px;box-sizing:border-box;margin:0;width:100%}.mw-parser-output table.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{vertical-align:top}.mw-parser-output table.commons-file-information-table>tbody>tr>td,.mw-parser-output table.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:4px}.mw-parser-output.fileinfo-paramfield{background:#ccf;text-align:right;padding-right:0.4em;width:15%;font-weight:bold}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{border-top:0;padding-top:0;margin-top:-8px}@media only screen and (max-width:719px){.mw-parser-output table.commons-file-information-table,.mw-parser-output.commons-file-information-table.fileinfotpl-type-information{border-spacing:0;padding:0;word-break:break-word;width:100%!important}.mw-parser-output.commons-file-information-table>tbody,.mw-parser-output.fileinfotpl-type-information>tbody{display:block}.mw-parser-output.commons-file-information-table>tbody>tr>td,.mw-parser-output.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:0.2em 0.4em;text-align:left;text-align:start}.mw-parser-output.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{display:flex;flex-direction:column}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{margin-top:-1px}.mw-parser-output.fileinfo-paramfield{box-sizing:border-box;flex:1 0 100%;width:100%}} Description: Deutsch: Genom von Thogotovirus English: Thogotovirus genome map. Date: (not specified). Source:
https://viralzone.expasy.org/resources/Thogotovirus%5Fgenomes.jpg. Author:
ViralZone, SIB
Swiss Institute of Bioinformatics:
Thogotovirus. Permission(
Reusing this file): : This work is
free and may be used by anyone for any purpose. If you wish to
use this content, you do not need to request permission as long as you follow any licensing requirements mentioned on this page. The Wikimedia Foundation has received an e-mail confirming that the copyright holder has approved publication under the terms mentioned on this page. This correspondence has been reviewed by a
Volunteer Response Team (VRT) member and stored in our
permission archive. The correspondence is available to trusted volunteers as
ticket #2021020910004221. If you have questions about the archived correspondence, please use the
VRT noticeboard. Ticket link:
https://ticket.wikimedia.org/otrs/index.pl?Action=AgentTicketZoom&TicketNumber=2021020910004221 Find other files from the same ticket:
.
-
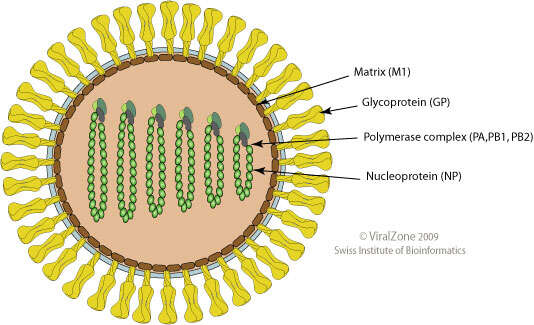
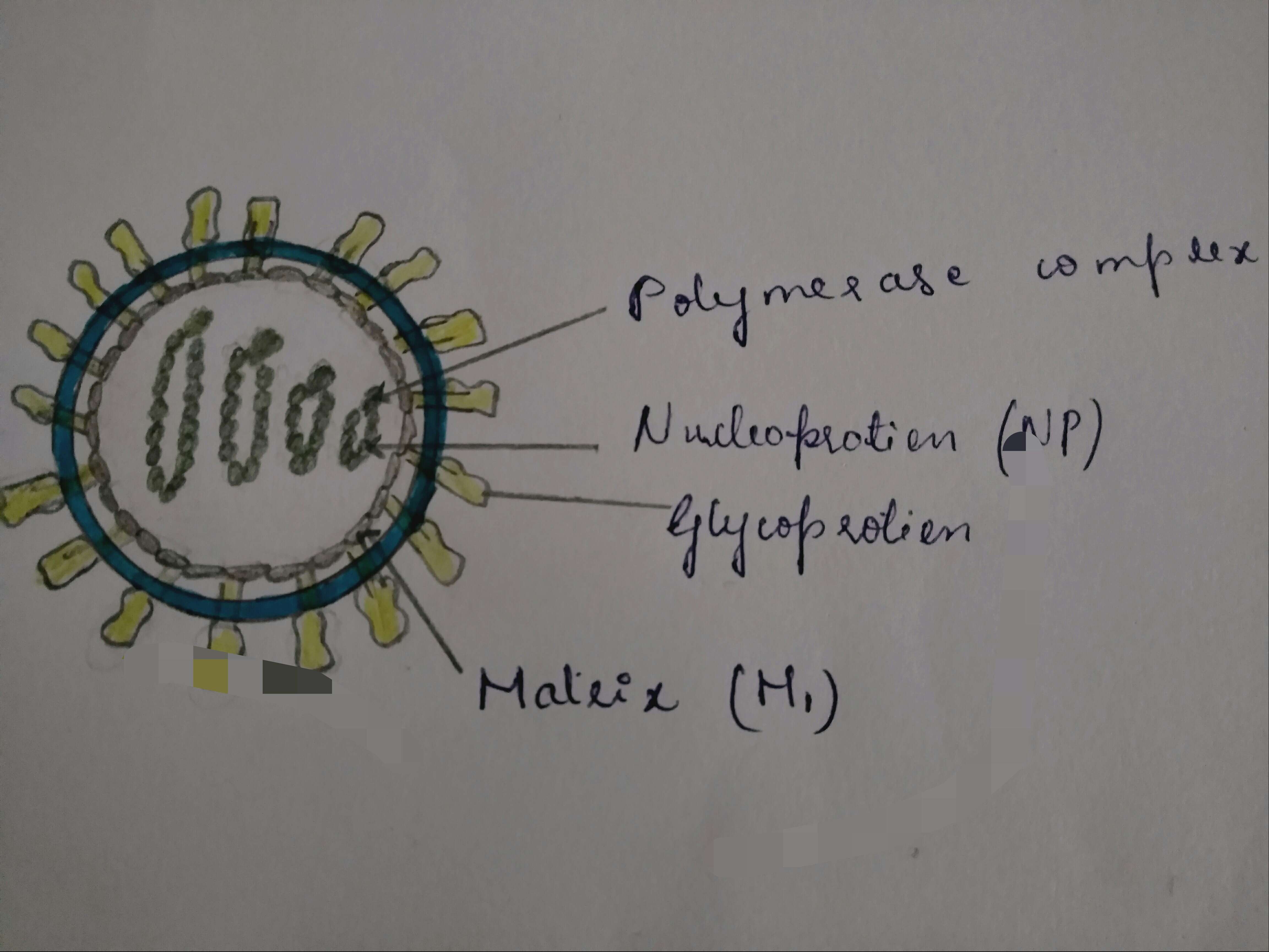
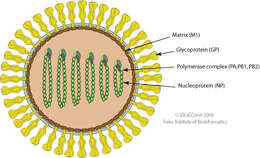
Summary.mw-parser-output table.commons-file-information-table,.mw-parser-output.fileinfotpl-type-information{border:1px solid #a2a9b1;background-color:#f8f9fa;padding:5px;font-size:95%;border-spacing:2px;box-sizing:border-box;margin:0;width:100%}.mw-parser-output table.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{vertical-align:top}.mw-parser-output table.commons-file-information-table>tbody>tr>td,.mw-parser-output table.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:4px}.mw-parser-output.fileinfo-paramfield{background:#ccf;text-align:right;padding-right:0.4em;width:15%;font-weight:bold}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{border-top:0;padding-top:0;margin-top:-8px}@media only screen and (max-width:719px){.mw-parser-output table.commons-file-information-table,.mw-parser-output.commons-file-information-table.fileinfotpl-type-information{border-spacing:0;padding:0;word-break:break-word;width:100%!important}.mw-parser-output.commons-file-information-table>tbody,.mw-parser-output.fileinfotpl-type-information>tbody{display:block}.mw-parser-output.commons-file-information-table>tbody>tr>td,.mw-parser-output.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:0.2em 0.4em;text-align:left;text-align:start}.mw-parser-output.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{display:flex;flex-direction:column}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{margin-top:-1px}.mw-parser-output.fileinfo-paramfield{box-sizing:border-box;flex:1 0 100%;width:100%}} Description: Deutsch: Schemazeichnung eines Virusteilchens (Gattungen Thogotovirus und Quaranjavirus, Querschnitt) English: Schematic drawing of a virion (Genera Thogotovirus and Quaranjavirus, cross section). Date: 2009. Source:
https://viralzone.expasy.org/resources/Thogotovirus%2Dvirion.jpg. Author:
ViralZone, SIB
Swiss Institute of Bioinformatics:
Quaranjavirus. Permission(
Reusing this file): : This work is
free and may be used by anyone for any purpose. If you wish to
use this content, you do not need to request permission as long as you follow any licensing requirements mentioned on this page. The Wikimedia Foundation has received an e-mail confirming that the copyright holder has approved publication under the terms mentioned on this page. This correspondence has been reviewed by a
Volunteer Response Team (VRT) member and stored in our
permission archive. The correspondence is available to trusted volunteers as
ticket #2021020910004221. If you have questions about the archived correspondence, please use the
VRT noticeboard. Ticket link:
https://ticket.wikimedia.org/otrs/index.pl?Action=AgentTicketZoom&TicketNumber=2021020910004221 Find other files from the same ticket:
.
-
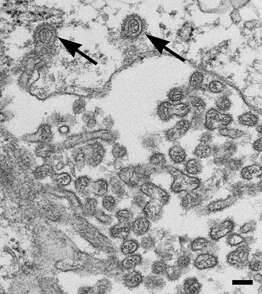
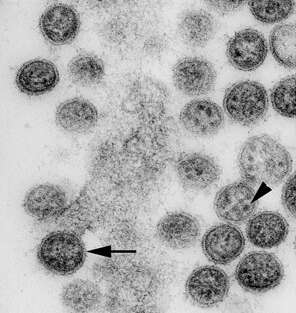
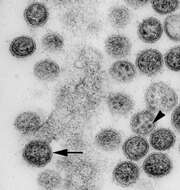
Description: Transmission electron micrograph of an ultrathin section of Vero cells infected with Cygnet River virus (CyRV) from a Muscovy duck, Australia. Arrow, virus budding from the plasma membrane; arrowhead, sand-like structures. Cropped from Figure 1a of Kessell et al. (2012) Cygnet River Virus, a Novel Orthomyxovirus from Ducks, Australia. Emerg. Infect. Dis. 18 doi: 10.3201/eid1812.120500. Date: 2012. Source:
http://wwwnc.cdc.gov/eid/article/18/12/12-0500-f1. Author: Allan Kessell, Alex Hyatt, Debra Lehmann, Songhua Shan, Sandra Crameri, Clare Holmes, Glenn Marsh, Catherine Williams, Mary Tachedjian, Meng Yu, John Bingham, Jean Payne, Sue Lowther, Jianning Wang, Lin-Fa Wang, and Ina Smith.
-
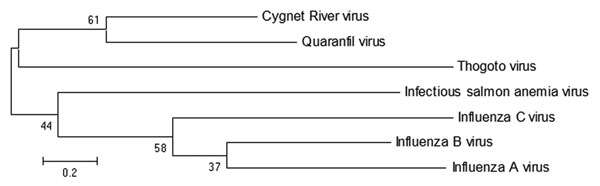
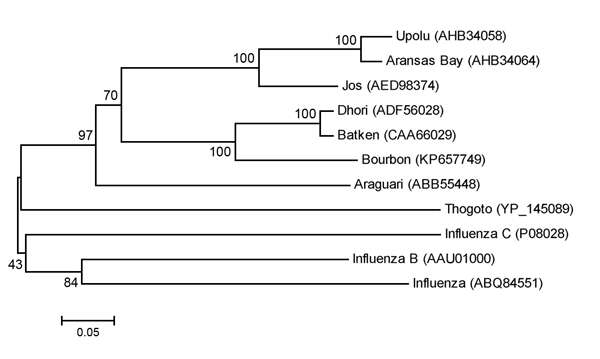
Description: English: Maximum-likelihood tree showing phylogenetic relationships between Cygnet River virus isolate 10–01646 (GenBank accession no. JQ693418) and other orthomyxoviruses: Quaranfil virus isolate EG T 377 (accession no. GQ499304), Thogoto virus strain PoTi503 (accession no. AF527530), infectious salmon anemia virus isolate RPC/NB (accession no. AF435424), influenza C virus C/Yamagata/8/96 (accession no. AB064433), influenza B virus B/Wisconsin/01/2010 (accession no. CY115184), and influenza A virus A/California/07/2009(H1N1) (accession no. CY121681). Tree was based on deduced amino acid sequences of the complete matrix protein of orthomyxoviruses, applying 1,000 bootstrap replicates (6). Numbers at nodes indicate percentage of 1,000 bootstrap replicates. Scale bar indicates nucleotide substitutions per site. Figure 2 of Kessell et al. (2012) Cygnet River Virus, a Novel Orthomyxovirus from Ducks, Australia. Emerg. Infect. Dis. 18 doi: 10.3201/eid1812.120500. Date: 2012. Source: wwwnc.cdc.gov/eid/article/18/12/12-0500-f2. Author: Allan Kessell, Alex Hyatt, Debra Lehmann, Songhua Shan, Sandra Crameri, Clare Holmes, Glenn Marsh, Catherine Williams, Mary Tachedjian, Meng Yu, John Bingham, Jean Payne, Sue Lowther, Jianning Wang, Lin-Fa Wang, and Ina Smith.
-
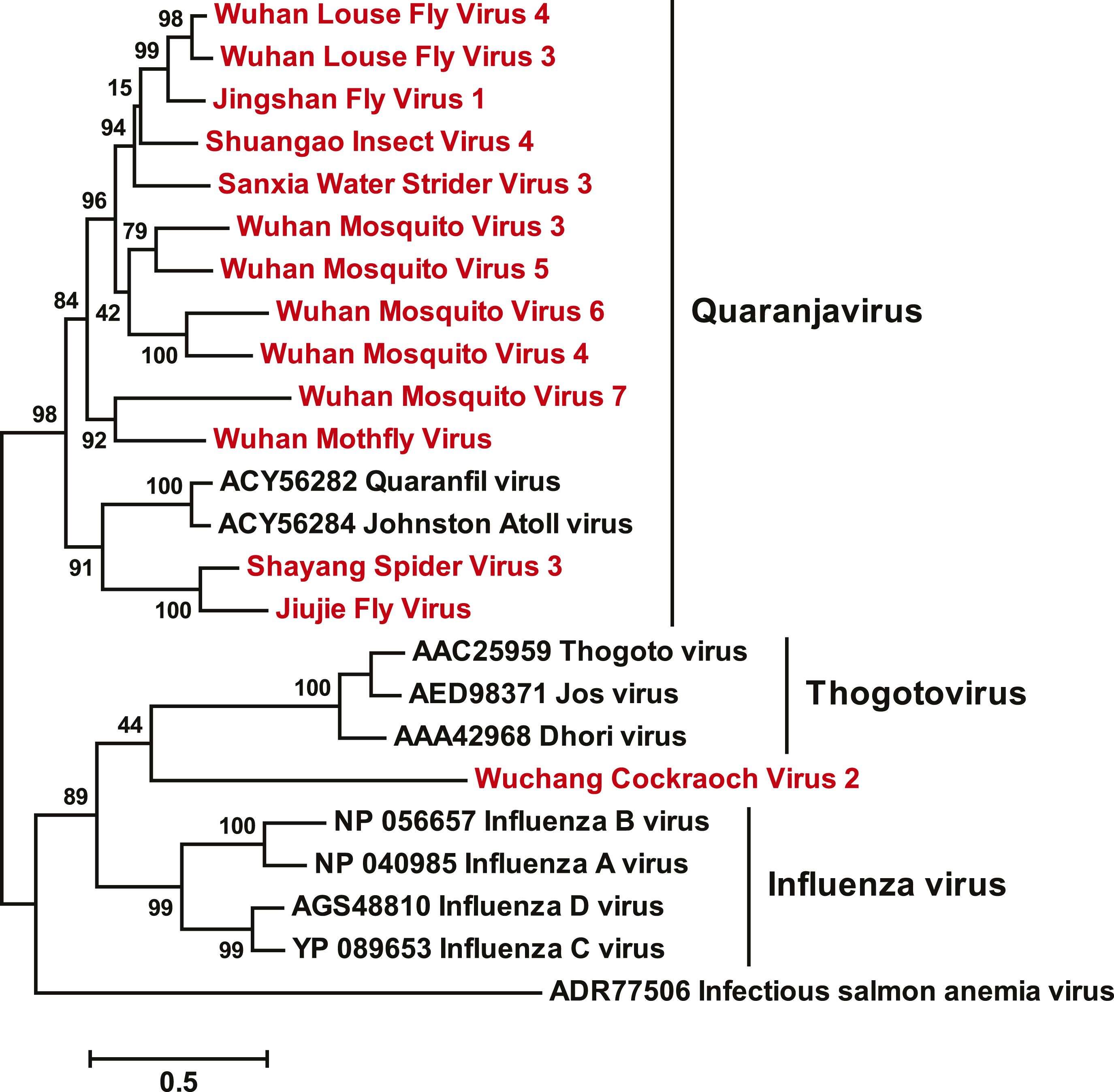
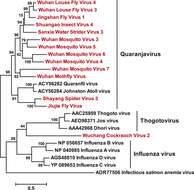
Description: English: ML phylogeny for the Orthomyxoviridae-like viruses. The phylogeny is reconstructed using RdRp alignments. Statistical support from the approximate likelihood-ratio test (aLRT) is shown on each node of the tree. The names of the viruses discovered in this study are shown in red. The names of reference sequences, which contain both the GenBank accession number and the virus species name, are shown in black. The names of previously defined genera/families are shown to the right of the phylogeny. Fig 3 suppl 1from Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, et al. (2015), "Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses", eLife 4: e05378. Date: 2015. Source:
http://elifesciences.org/content/elife/4/e05378/F3/F4.large.jpg. Author: Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, et al.
-
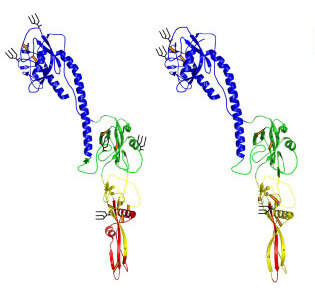
Description: English: Models of THOV GP (left) and AcMNPV GP64 (right) based on the X-ray crystallographic structure of VSV G. Key: blue: domain III; green: domain I; yellow: domain II; red: fusion loops; orange/black lines: dicysteine linkages; black lines: N-glycosylation sites. Date: 2008. Source:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2288602/. Author: Courtney E Garry and Robert F Garry.
-
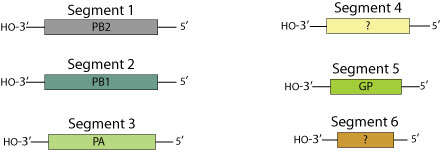
Summary.mw-parser-output table.commons-file-information-table,.mw-parser-output.fileinfotpl-type-information{border:1px solid #a2a9b1;background-color:#f8f9fa;padding:5px;font-size:95%;border-spacing:2px;box-sizing:border-box;margin:0;width:100%}.mw-parser-output table.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{vertical-align:top}.mw-parser-output table.commons-file-information-table>tbody>tr>td,.mw-parser-output table.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:4px}.mw-parser-output.fileinfo-paramfield{background:#ccf;text-align:right;padding-right:0.4em;width:15%;font-weight:bold}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{border-top:0;padding-top:0;margin-top:-8px}@media only screen and (max-width:719px){.mw-parser-output table.commons-file-information-table,.mw-parser-output.commons-file-information-table.fileinfotpl-type-information{border-spacing:0;padding:0;word-break:break-word;width:100%!important}.mw-parser-output.commons-file-information-table>tbody,.mw-parser-output.fileinfotpl-type-information>tbody{display:block}.mw-parser-output.commons-file-information-table>tbody>tr>td,.mw-parser-output.commons-file-information-table>tbody>tr>th,.mw-parser-output.fileinfotpl-type-information>tbody>tr>td,.mw-parser-output.fileinfotpl-type-information>tbody>tr>th{padding:0.2em 0.4em;text-align:left;text-align:start}.mw-parser-output.commons-file-information-table>tbody>tr,.mw-parser-output.fileinfotpl-type-information>tbody>tr{display:flex;flex-direction:column}.mw-parser-output.commons-file-information-table+table.commons-file-information-table,.mw-parser-output.commons-file-information-table+div.commons-file-information-table>table{margin-top:-1px}.mw-parser-output.fileinfo-paramfield{box-sizing:border-box;flex:1 0 100%;width:100%}} Description: Deutsch: Genom von Quaranjavirus English: Quaranjavirus genome map. Date: (not specified). Source:
https://viralzone.expasy.org/resources/Quaranjavirus%5Fgenomes.jpg. Author:
ViralZone, SIB
Swiss Institute of Bioinformatics:
Quaranjavirus. Permission(
Reusing this file): : This work is
free and may be used by anyone for any purpose. If you wish to
use this content, you do not need to request permission as long as you follow any licensing requirements mentioned on this page. The Wikimedia Foundation has received an e-mail confirming that the copyright holder has approved publication under the terms mentioned on this page. This correspondence has been reviewed by a
Volunteer Response Team (VRT) member and stored in our
permission archive. The correspondence is available to trusted volunteers as
ticket #2021020910004221. If you have questions about the archived correspondence, please use the
VRT noticeboard. Ticket link:
https://ticket.wikimedia.org/otrs/index.pl?Action=AgentTicketZoom&TicketNumber=2021020910004221 Find other files from the same ticket:
.
-
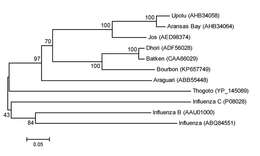
Description: English: Phylogeny of deduced amino acid sequences of Bourbon virus in comparison to homologous sequences of selected orthomyxoviruses. A neighbor-joining method was used for inference of each phylogeny with 2,000 replicates for bootstrap testing. Values at nodes are bootstrap values. Based on nucleocapsid protein (segment 5). Scale bar indicates number of amino acid substitutions per site. Date: 2015. Source:
http://wwwnc.cdc.gov/eid/article/21/5/15-0150-f2. Author: Olga I. Kosoy, Amy J. Lambert, Dana J. Hawkinson, Daniel M. Pastula, Cynthia S. Goldsmith, D. Charles Hunt, and J. Erin Staples.
-
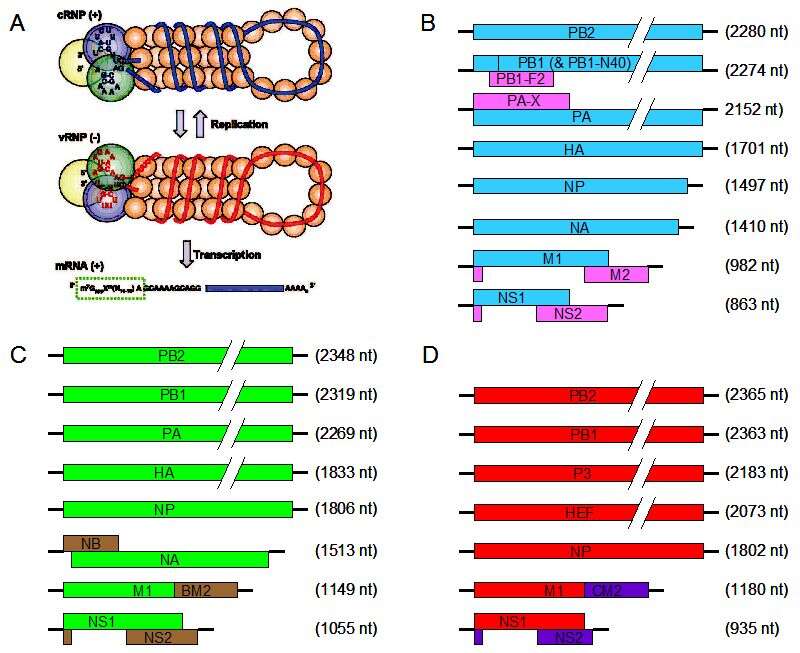
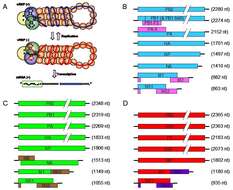
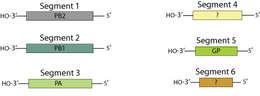
Description: English: Genomes of influenza viruses. A) The ends of the negative strand influenza genomic RNA are complexed with the three polymerase proteins and the remaining sequence is encapsidated with nucleoprotein (vRNP (-)). The positive strand cRNP is similarly complexed. The mRNA is transcribed with a 5’ cap structure and poly-A tail (see main text for details). Figure used with permission from Resa-Infante et al., 2011. B) Schematic of the influenza A virus genome. The bold black lines represent the 3’ and 5’ untranslated regions. The blue and pink boxes represent the major protein coding regions. C) Schematic of the influenza B virus genome. The green and brown boxes represent the major protein coding regions. D) Schematic of the influenza C virus genome. The red and purple boxes represent the major protein coding regions. The protein coding regions are not to scale. Coding regions in a different reading frame are shown above or below each other, coding regions in the same frame are show as contiguous blocks. Date: 20 November 2013. Source:
https://www.intechopen.com/books/current-issues-in-molecular-virology-viral-genetics-and-biotechnological-applications/gene-constellation-of-influenza-vaccine-seed-viruses. Author: Ewan P. Plant and Zhiping Ye.
-
imageDan Dou, Rebecca Revol, Henrik Östbye, Hao Wang, and Robert Daniels
Wikimedia Commons
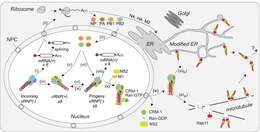
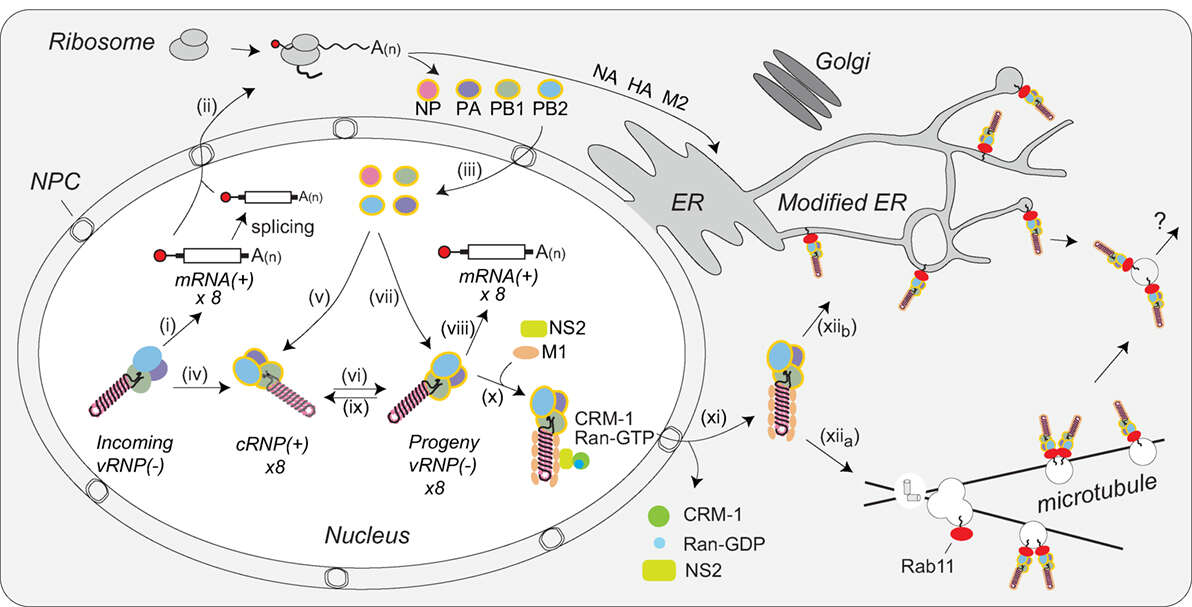
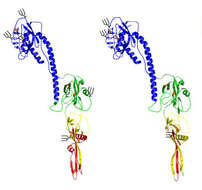
Description: English: Coordination of viral ribonucleoprotein (vRNP) assembly and trafficking to the plasma membrane. Upon entry into the host cell nucleus, (i) the vRNP-associated viral polymerase transcribes the viral mRNAs. (ii) The mRNAs are either directly, or after alternative splicing, exported for translation by cytosolic ribosomes. (iii) Newly synthesized viral polymerase subunits (PA, PB1, and PB2) and nucleoprotein (NP) are imported back into the nucleus. (iv) Due to the inefficient dinucleotide priming, the vRNP-associated viral polymerase also infrequently transcribes complimentary RNA (cRNA) copies that assemble into cRNPs via (v) binding of a newly synthesized viral polymerase (PA, PB1, and PB2) and NP. (vi) The polymerase transcribes viral RNA (vRNA) copies from the positive strand in the cRNPs and these assemble into vRNPs by (vii) association with a new viral polymerase (PA, PB1, and PB2) and NP. Once assembled, the new vRNPs can (viii) transcribe additional viral mRNAs, (ix) transcribe new cRNA copies, or (x) associate with the newly synthesized viral proteins M1 and NS2 to facilitate the recruitment of CRM1, which (xi) mediates the nuclear export of the vRNP. (xiia) Once exported, the vRNPs then associate with Rab11 that assists in the trafficking of the vRNPs toward the cell surface. The vRNP trafficking either occurs by Rab11-containing vesicles associated with microtubules or (xiib) through Rab11 located in the modified endoplasmic reticulum (ER) membranes. How the vRNPs reach the budding site at the plasma membrane is currently not known. Date: 20 July 2018. Source:
https://www.frontiersin.org/articles/10.3389/fimmu.2018.01581/full. Author: imageDan Dou, Rebecca Revol, Henrik Östbye, Hao Wang, and Robert Daniels.
-
Description: English: Transmission electron micrograph of influenza A virus, late passage. Deutsch: aviäres Influenzavirus (HPAIV), elektronenmikroskopische Aufnahme 日本語: 高病原性トリインフルエンザウイルス, 長期間継代培養したトリインフルエンザウイルスの透過電子顕微鏡像。 한국어: 조류 독감을 일으키는 인플루엔자바이러스 A형. Date: 1981. Source: : This media comes from the
Centers for Disease Control and Prevention's
Public Health Image Library (PHIL), with identification number
#280. Note: Not all PHIL images are public domain; be sure to check copyright status and credit authors and content providers.
العربية |
Deutsch |
English |
македонски |
slovenščina |
+/−. Author: Photo Credit: Content Providers(s): CDC/ Dr. Erskine Palmer. Permission(
Reusing this file): PD-USGov-HHS-CDC (None - This image is in the public domain and thus free of any copyright restrictions. As a matter of courtesy we request that the content provider be credited and notified in any public or private usage of this image.).
-
Description: Español: Mauro Zernotti, Médico Clínico, Sobre los grupos de riesgo de la Gripe A (H1N1). Date: 2 June 2016. Source: Own work. Author:
María Antonieta Cacciavillani.
-
Description: Colorized transmission electron micrograph of negatively stained SW31 (swine strain) influenza virus particles. Credit: NIAID. Date: 29 July 2014, 11:56. Source:
Swine Flu Strain Virus Particles. Author:
NIAID.