: All images in this gallery could be recreated using
vector graphics as an
SVG file. This has several advantages; see
Commons:Media for cleanup for more information. If an SVG form of this image is available, please upload it and afterwards replace this template with {{
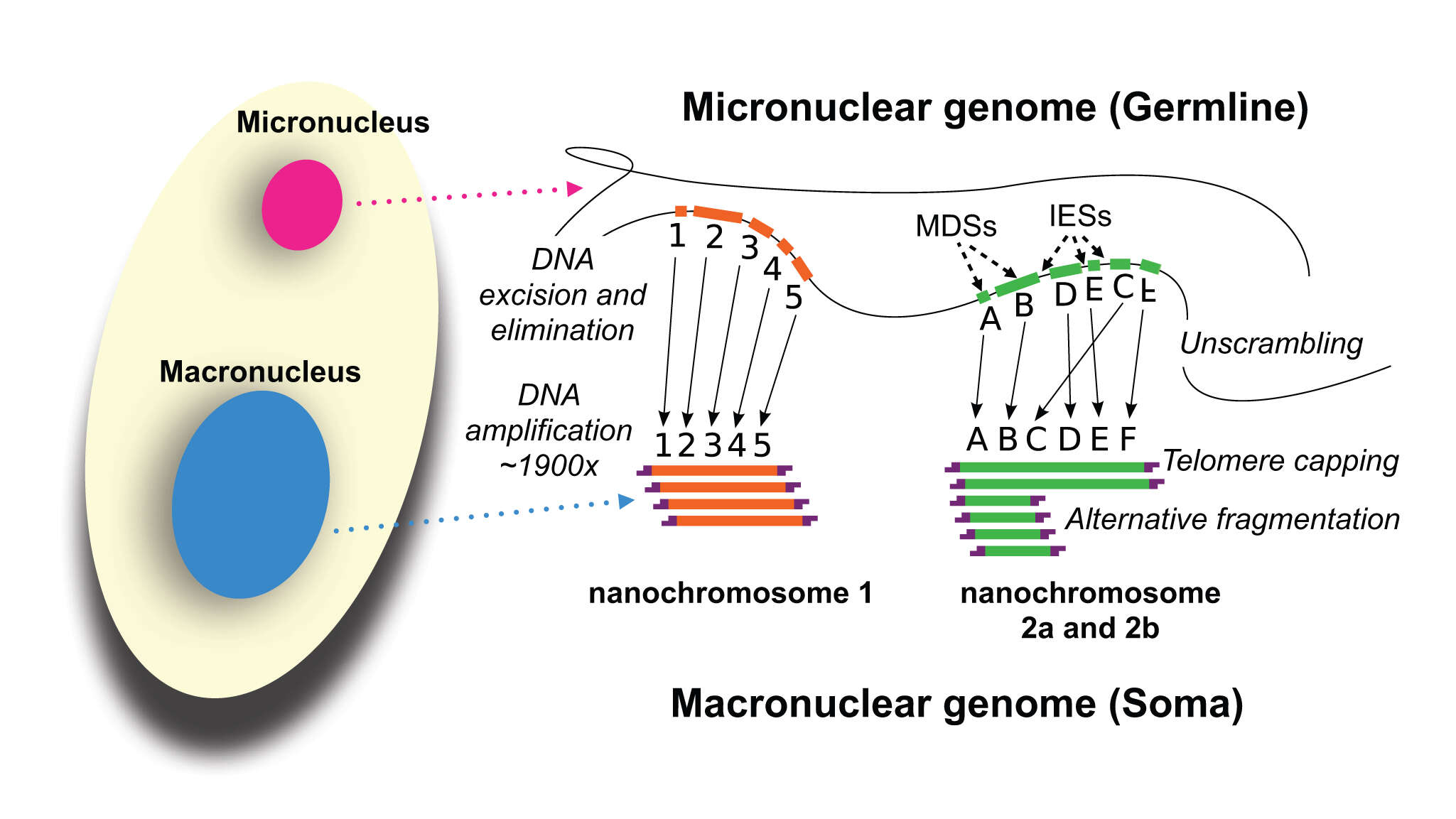
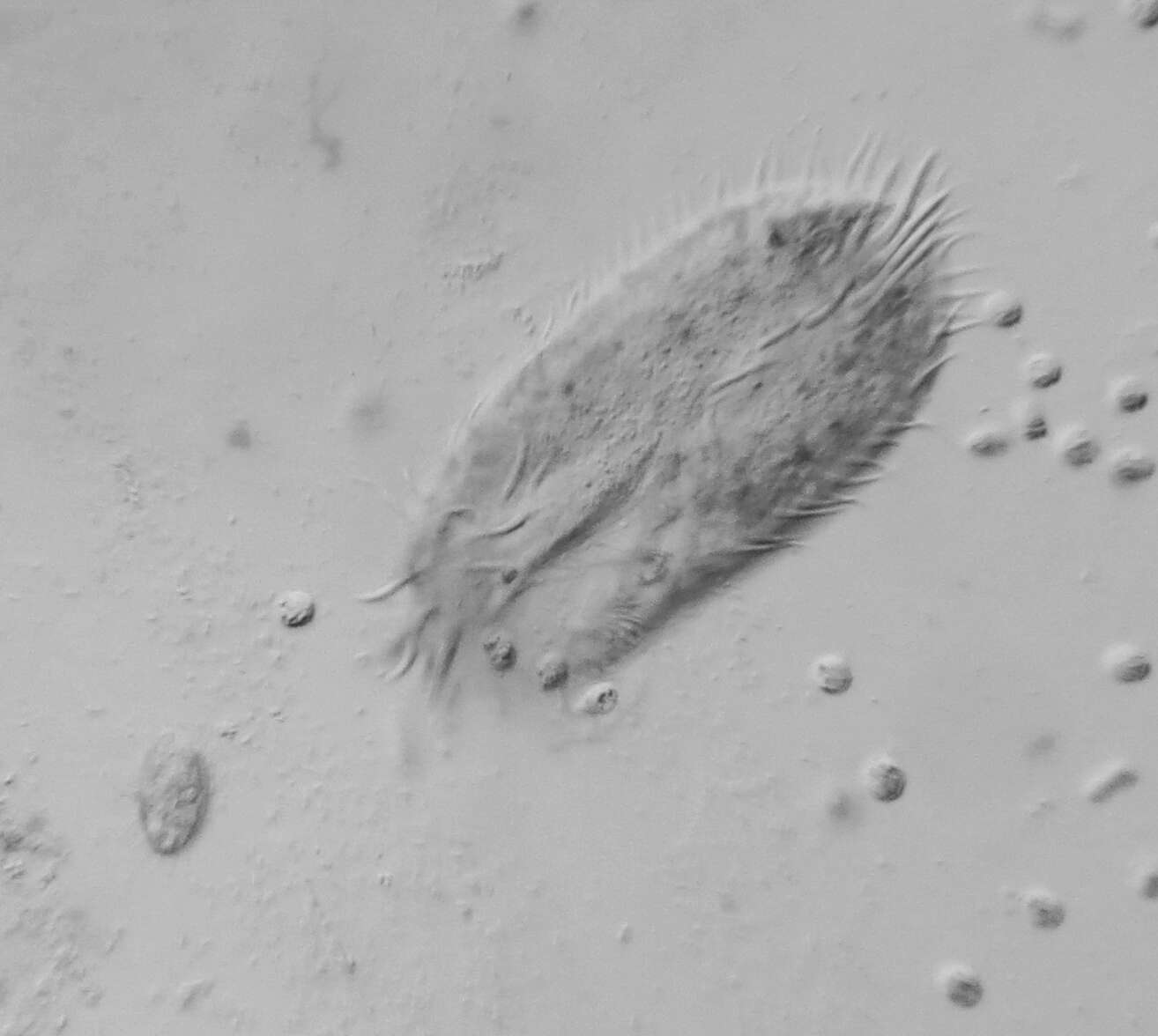
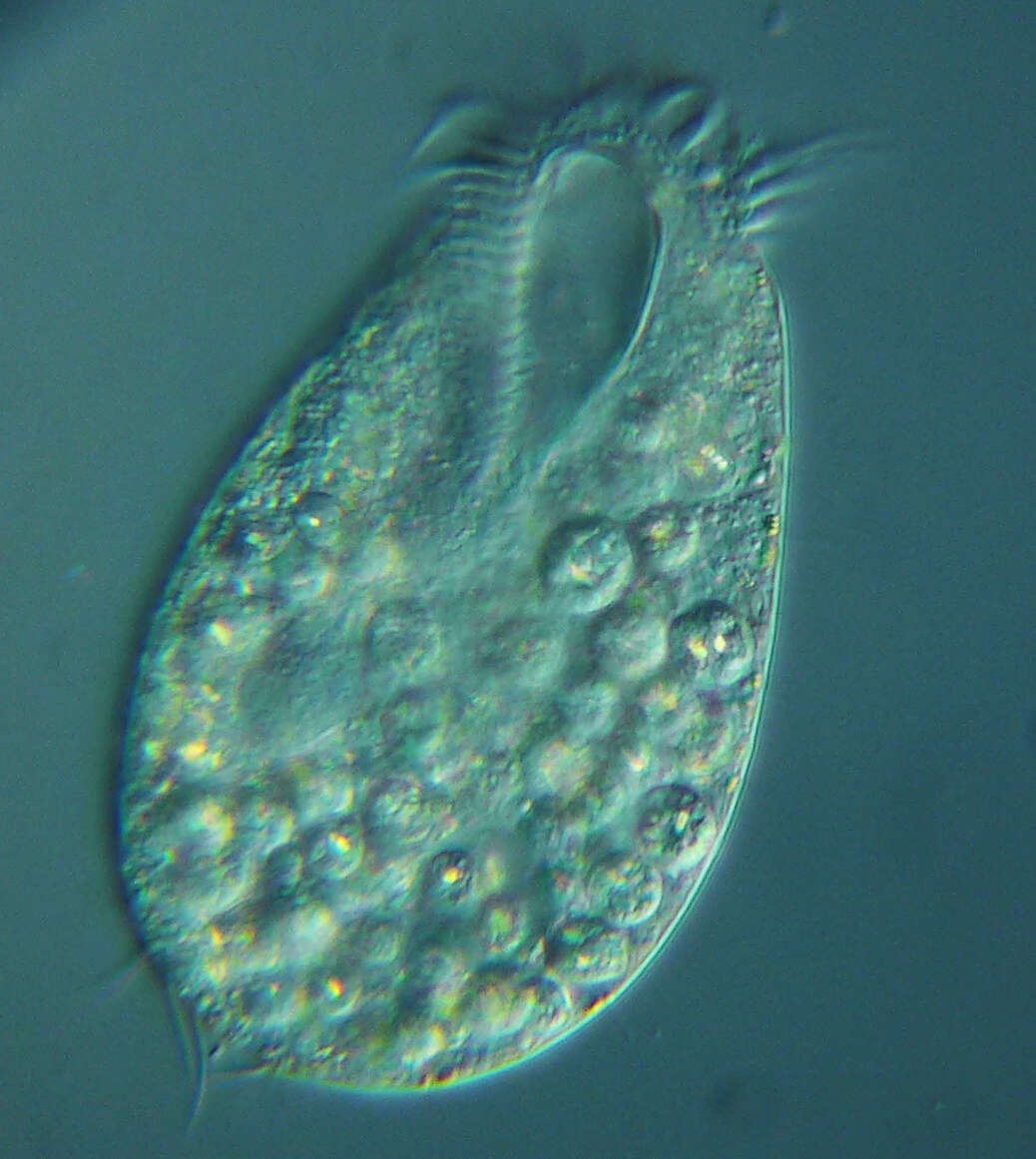
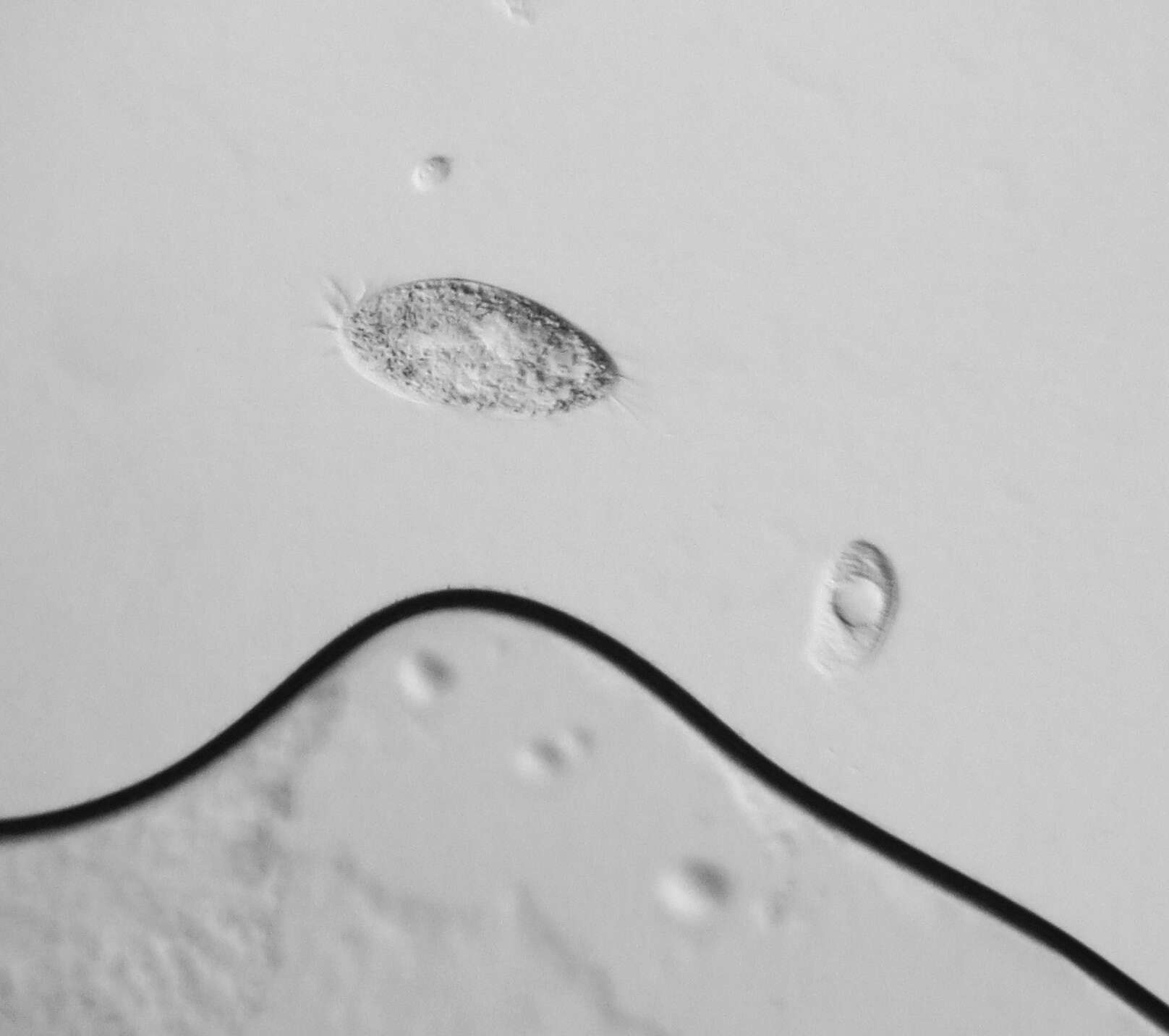
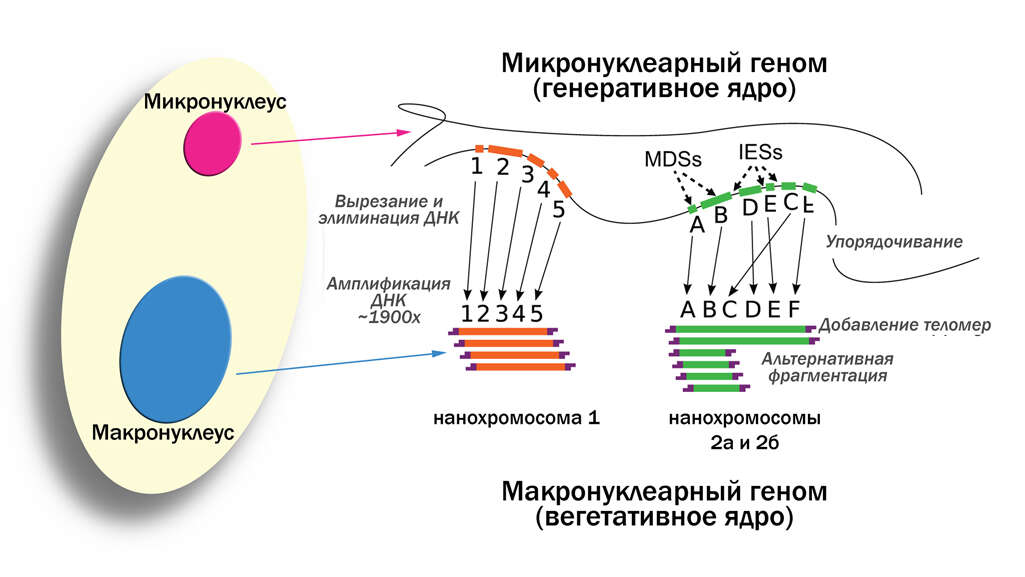
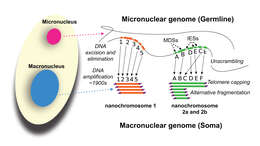
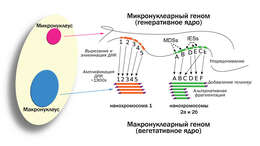
vector version available|new image name}}. Description: English: Development of the Oxytricha macronuclear genome from the micronuclear genome: During conjugation of Oxytricha cells, segments of the micronuclear genome (MDSs) are excised and stitched together to form the nanochromosomes of the new macronuclear genome, and the remainder of the micronuclear genome is eliminated (including the IESs interspersed between MDSs). The old macronuclear genome is also degraded during development. The segments that are stitched together may be either in order (e.g., forming nanochromosome 1, on the left) or out of order or inverted (e.g., forming the two forms of nanochromosome 2), in which case they need to be “unscrambled.” Two rounds of DNA amplification produce nanochromosomes at an average copy number of ~1,900. Alternative fragmentation of DNA during nanochromosome development may also occur, irrespective of unscrambling, giving rise to longer (2a) and shorter (2b) nanochromosome isoforms. The mature nanochromosomes are capped on both ends with telomeres. Date: 27 February 2013, 23:42:45. Source: Swart EC, Bracht JR, Magrini V, Minx P, Chen X, et al. (2013) The Oxytricha trifallax Macronuclear Genome: A Complex Eukaryotic Genome with 16,000 Tiny Chromosomes. PLoS Biol 11(1): e1001473. doi:10.1371/journal.pbio.1001473. Author: Swart et al. Other versions: Derivative works of this file:
Oxytricha Ma genome - rus.jpg.