-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
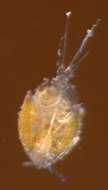
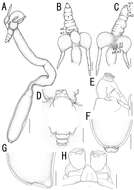
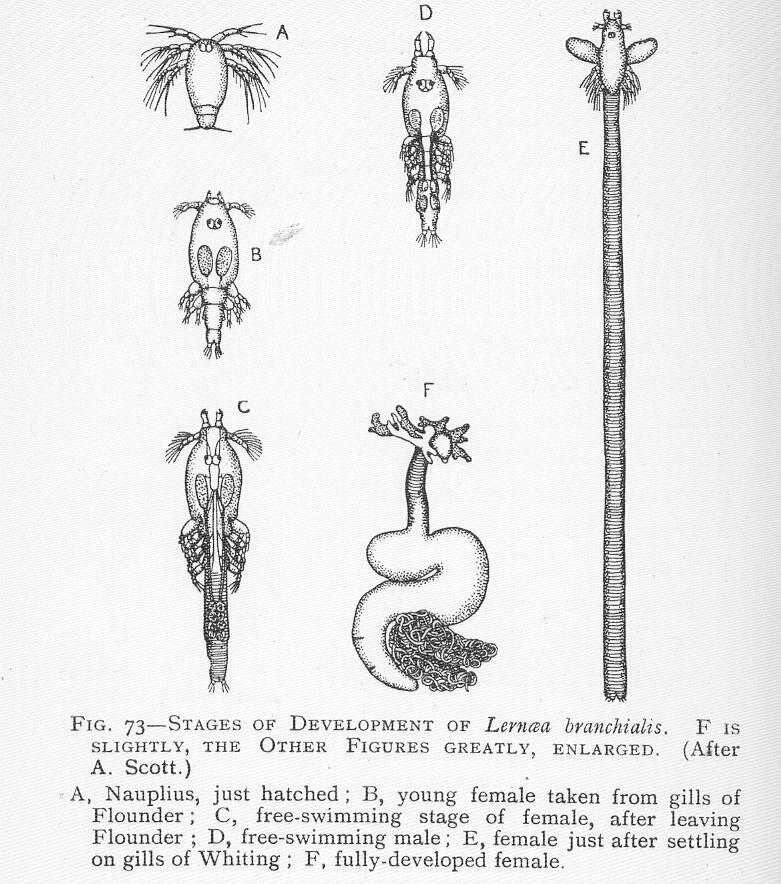
Stages of development of Lernaea branchialis. A, Nauplius, just hatched; B, young female taken from gills of flounder; C, free-swimming stage of female, after leaving flounder; D, free-swimming male; E, female just after settling on gills of whiting; F, fully-developed female
-
Eduardo Suárez-Morales, Humberto Camisotti, Alberto Martín
Zookeys
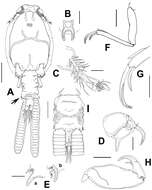
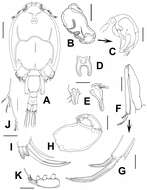
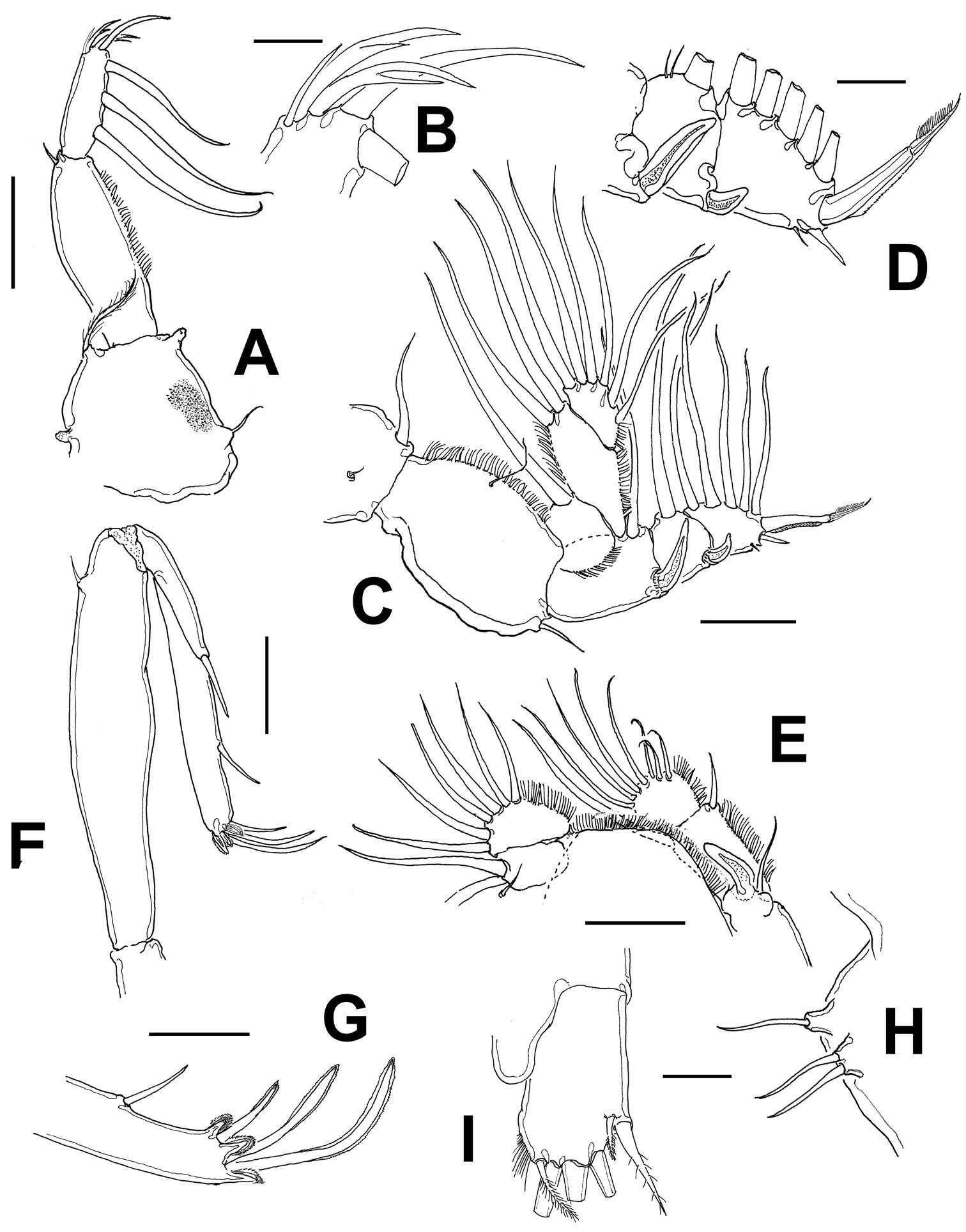
Figure 1.Caligus evelynae sp.n., adult female from Venezuela: A habitus, dorsal view B sternal furca, ventral view C antennule D antenna E postantennal process (b) and maxillule (a) F maxilla G detail of calamus and canna H maxilliped I genital complex and abdomen, ventral view. Scale bars: A, I=0.5 mm, B–F, H =0.1 mm, G=0.05 mm.
-
Daisuke Uyeno, Kaori Wakabayashi, Kazuya Nagasawa
Zookeys
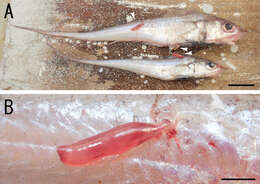
Figure 1.Sarcotretes umitakae sp. n., female on Coelorinchus jordani Smith and Pope. A two specimens of Coelorinchus jordani (181.5 mm TL and 142.8 mm TL) carrying the type series of Sarcotretes sp. n. (arrowheads) B coloration in life of paratype NSMT–Cr 22254 attached to host’s body. Scale bars: A=20 mm; B=3 mm.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
Eduardo Suárez-Morales, Humberto Camisotti, Alberto Martín
Zookeys
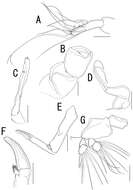
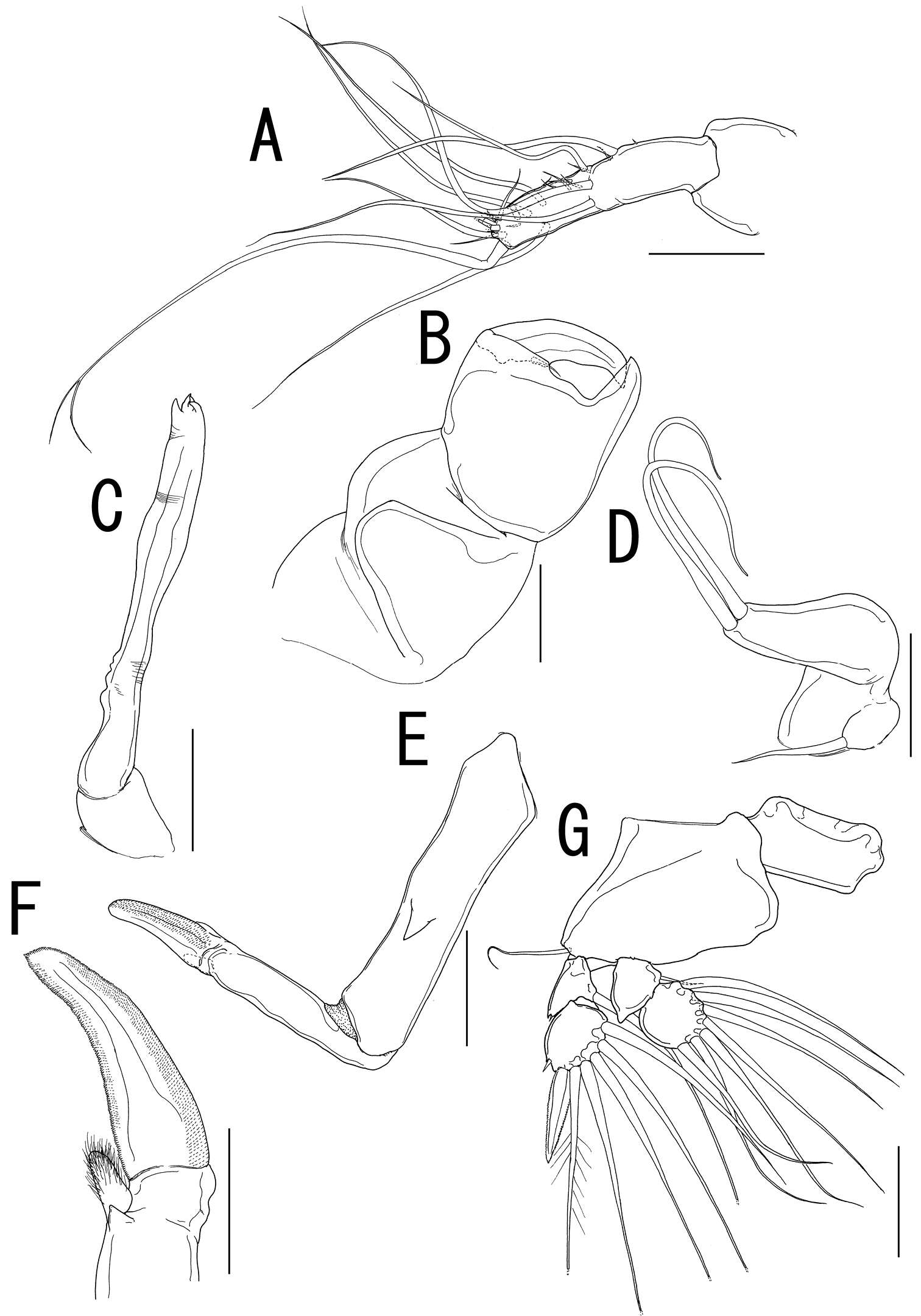
Figure 2.Caligus evelynae sp.n., adult female from Venezuela: A first leg B detail of distal elements of first leg C second leg D detail of exopodal segments of second leg E third leg F fourth leg G detail of terminal elements of fourth leg H fifth leg I caudal ramus, dorsal. Scale bars: A,C, F, E=0.1 mm, B, D, G, H, I=0.025 mm.
-
Daisuke Uyeno, Kaori Wakabayashi, Kazuya Nagasawa
Zookeys
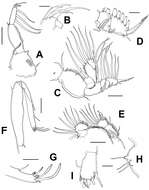
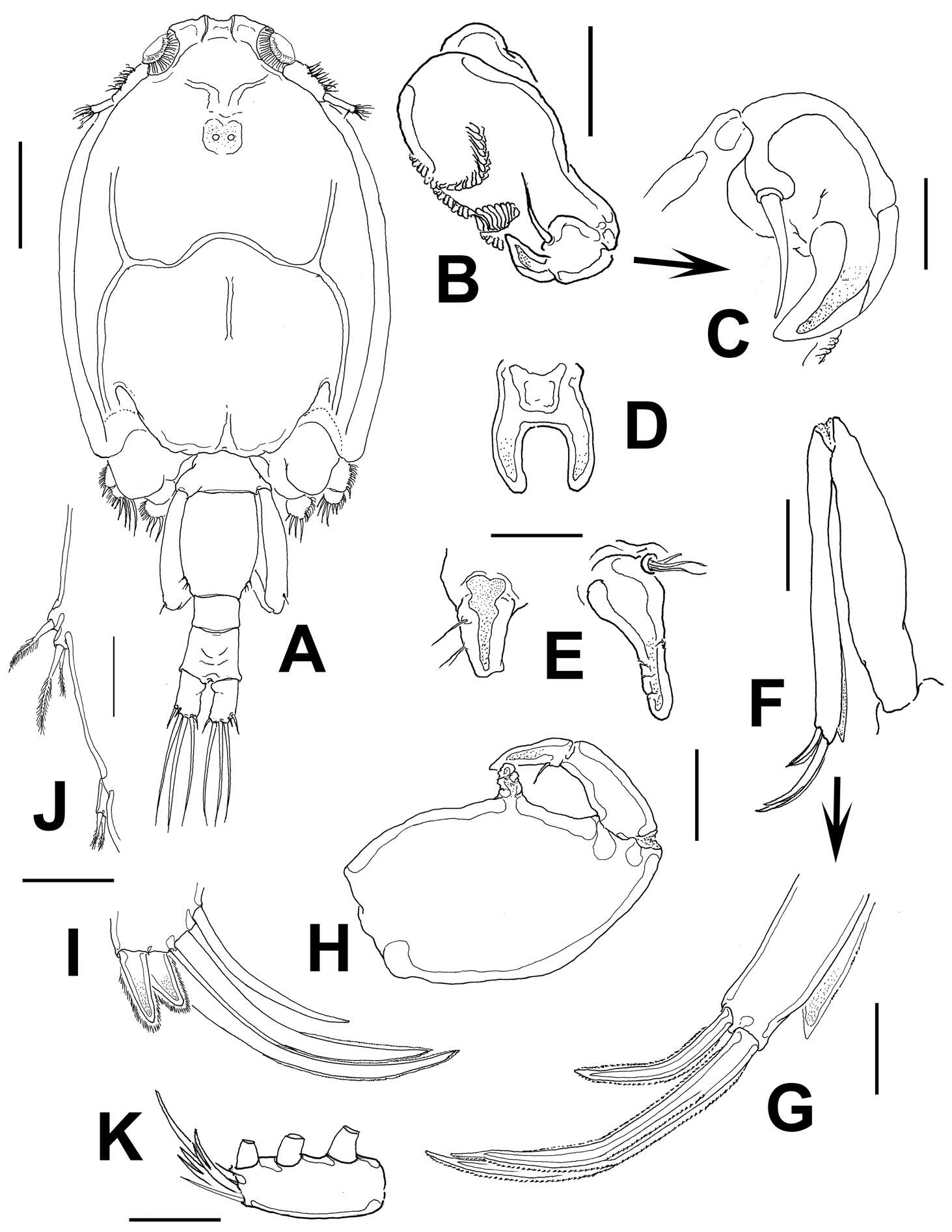
Figure 2.Sarcotretes umitakae sp. n., female, holotype NSMT–Cr 22253. A habitus B anterior portion of body, dorsal, a1 = antennule, a2 = antenna C same, ventral, m1 = maxillule, m2 = maxilla, p1 = leg 1, p2 = leg 2, p3 = leg 3, p4 = vestige of leg 4 D vestige of dorsal cephalothoracic shield E tip of proboscis, lateral F posterior portion of body, ventral G same, lateral H rostral area and antennae, dorsal. Scale bars: A=3 mm; B, C, F, G=1 mm; D=500 μm; E=300 μm; H=150 μm.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
Eduardo Suárez-Morales, Humberto Camisotti, Alberto Martín
Zookeys
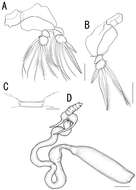
Figure 3.Caligus evelynae sp.n., adult male from Venezuela: A habitus, dorsal view B antenna C detail of distal part of antenna D sternal furca, ventral view E postantennal process and maxillule F maxilla G detail of calamus and canna H maxilliped I fourth leg, detail of distal elements J fifth and sixth legs K first leg, distal segment of exopod. Scale bars: A=0.5 mm, B,D–F, H=0.1 mm, C, G, I, J=0.03 mm; K=0.07 mm.
-
Daisuke Uyeno, Kaori Wakabayashi, Kazuya Nagasawa
Zookeys
Figure 3.Sarcotretes umitakae sp. n., female, holotype NSMT–Cr 22253. A left antennule, anterior B left antenna, anterior C left mandible D left maxillule E left maxilla, lateral F distal part of left maxilla G right leg 1, anterior. Scale bars: A, B, E, G=100 μm; C, D=70 μm; F=50μm.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
Daisuke Uyeno, Kaori Wakabayashi, Kazuya Nagasawa
Zookeys
Figure 4.Sarcotretes umitakae sp. n., female, holotype NSMT–Cr 22253. A left leg 2, anterior B right leg 3, anterior C vestige of leg 4. Sarcotretes umitakae sp. n., female, paratype NSMT–Cr 22254 D habitus. Scale bars: A, B=100 μm; C=30 μm; D=3 mm.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.