en
names in breadcrumbs

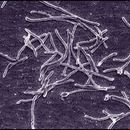
Thermus aquaticus was originally isolated from a number of hot springs in Yellowstone National Park and a hot spring in California (U.S.A.) and was subsequently isolated from hot springs in other parts of the world and even from artificial hot water environments such as hot tap water (Brock and Freeze 1969; Brock and Boylen 1973). Previously, microbiologists had enriched for thermophilic bacteria (i.e, bacteria that thrive at high temperatures) at 55° C, but the discoverer of T. aquaticus, Thomas Brock, found that many thermophiles in his studies of microbial ecology would not be easily detected at such a "low" temperature since they require temperatures above 70° C to flourish.
Gibbs et al. (2009) studied the phylogeny of Thermus isolates and found they fell into 8 clades. They then isolated the DNA polymerase I genes from 22 representatives and cloned the 8 most diverse genes (to represent the 8 clades) into an expression vector. They were able to purify the protein from six of these clones and examined their biochemical characteristics and suitability for PCR. They found that none was as thermostable as commercially available Taq polymerase and all had error-frequencies similar to those of Taq polymerase. They concluded that the initial selection of T. aquaticus for DNA polymerase purification was a fortuitous choice, although they suggest that simple mutagenesis procedures on other Thermus-derived polymerases should provide comparable thermostability for the PCR reaction.
Degryse et al. (1978) and Degryse and Glansdorff (1981) studied the metabolism of Thermus aquaticus. These bacteria are obligately aerobic and chemoheterotrophic (Degryse et al. 1978).
The use of DNA polymerases from T. aquaticus and other thermophiles in the polymerase chain reaction (PCR) and related applications, such as DNA sequencing, has revolutionized biotechnology. The humble T. aquaticus enormously expanded what questions could be practically addressed in fields ranging from biomedical science ("what is the genetic basis for disease X and does this patient have this disorder?") to animal behavior ("were all the young in this bluebird nest actually sired by the mother's apparent mate?") to conservation ("is this whale meat being sold truly from the species the seller claims?") to forensics ("can this accused criminal possibly have left the DNA evidence found at the crime scene?") and beyond.
In addition to its DNA polymerase, a range of other thermostable enzymes have been isolated from T. aquaticus for use in high temperature molecular biology applications. Largely as a result of the example of T. aquaticus, which has generated enormous profits for the biotechnology industry but not for the national park in which it was discovered (nor for U.S. taxpayers), the National Park Service now may require researchers working in national parks to sign "benefits sharing" agreements requiring that profits that may be derived at a later point in time from work done in the park be shared with the park.
Thermus aquaticus is a thermophilic gram-negative bacterium that has played a key role in the modern revolution in genetic research, genetic engineering, and biotechnology (see below). Thermophilic bacteria are bacteria that thrive at very high temperatures, often above 45° C (113° F). Thermus aquaticus was originally isolated from a number of hot springs in Yellowstone National Park and a hot spring in California (U.S.A.) and was subsequently isolated from hot springs in other parts of the world and even from artificial hot water environments such as hot tap water (Brock and Freeze 1969; Brock and Boylen 1973). Previously, microbiologists had enriched for thermophilic bacteria at 55° C, but the discoverer of T. aquaticus, Thomas Brock, found that many thermophiles in his studies of microbial ecology would not be easily detected at such a "low" temperature since they require temperatures above 70° C to flourish. As Brock has noted, his discovery provides an excellent illustration of the often unpredictable value (both academic and applied) of basic research (Brock 1997).
In the 1980s, a method known as the polymerase chain reaction (PCR) was developed to generate many copies of targeted segments of DNA from very tiny samples (Mullis and Faloona 1987; Arnheim and Erlich 1992; visit this site for a visual explanation of the principle of PCR) . This technique includes repeated cycles of melting apart of the two strands of each double-stranded DNA molecule (typically at 92° to 95° C) alternating with the extension of new complementary strands to create additional copies. To be practical, this method requires the use of a DNA polymerase that is not destroyed by this heating (DNA polymerases are enzymes that play a key role in DNA synthesis within cells; see Pavlov et al. 2004 for an overview of DNA polymerases). Fortunately, evolution has provided such polymerases in bacteria adapted to live at very high temperatures (e.g., in hot springs). Chien et al. (1976) had already purified a stable DNA polymerase from T. aquaticus with a temperature optimum of 80° C, which proved to serve very well for the automation of PCR.
The use of DNA polymerases from T. aquaticus and other thermophiles in PCR and related applications, such as DNA sequencing, has revolutionized biotechnology. The humble T. aquaticus enormously expanded what questions could be practically addressed in fields ranging from biomedical science ("what is the genetic basis for disease X and does this patient have this disorder?") to animal behavior ("were all the young in this bluebird nest actually sired by the mother's apparent mate?") to conservation ("is this whale meat being sold truly from the species the seller claims?") to forensics ("can this accused criminal possibly have left the DNA evidence found at the crime scene?") and beyond.
Thermus aquaticus is a species of bacteria that can tolerate high temperatures, one of several thermophilic bacteria that belong to the Deinococcota phylum. It is the source of the heat-resistant enzyme Taq DNA polymerase, one of the most important enzymes in molecular biology because of its use in the polymerase chain reaction (PCR) DNA amplification technique.
When studies of biological organisms in hot springs began in the 1960s, scientists thought that the life of thermophilic bacteria could not be sustained in temperatures above about 55 °C (131 °F).[1] Soon, however, it was discovered that many bacteria in different springs not only survived, but also thrived in higher temperatures. In 1969, Thomas D. Brock and Hudson Freeze of Indiana University reported a new species of thermophilic bacteria which they named Thermus aquaticus.[2] The bacterium was first isolated from Mushroom Spring in the Lower Geyser Basin of Yellowstone National Park, which is near the major Great Fountain Geyser and White Dome Geyser,[3] and has since been found in similar thermal habitats around the world.
T. aquaticus shows best growth at 65–70 °C (149–158 °F), but can survive at temperatures of 50–80 °C (122–176 °F). It primarily scavenges for protein from its environment as is evidenced by the large number of extracellular and intracellular proteases and peptidases as well as transport proteins for amino acids and oligopeptides across its cell membrane. This bacterium is a chemotroph—it performs chemosynthesis to obtain food. However, since its range of temperature overlaps somewhat with that of the photosynthetic cyanobacteria that share its ideal environment, it is sometimes found living jointly with its neighbors, obtaining energy for growth from their photosynthesis. T. aquaticus normally respires aerobically but one of its strains, Thermus aquaticus Y51MC23, is able to be grown anaerobically.[4]
The genetic material of T. aquaticus consists of one chromosome and four plasmids, and its complete genome sequencing revealed CRISPR genes at numerous loci.[5]
Thermus aquaticus is generally of cylindrical shape with a diameter of 0.5 μm to 0.8 μm. The shorter rod shape has a length of 5 μm to 10 μm. The longer filament shape has a length that varies greatly and in some cases exceeds 200 μm. T. aquaticus has shown multiple possible morphologies in different cultures. The rod-shaped bacteria have a tendency to aggregate. Associations of several individuals can lead to the formation of spherical bodies 10 μm to 20 μm in diameter, also called rotund bodies.[2][6] These bodies are not composed of cell envelope or outer membrane components as previously thought, but are instead made from remodelled peptidoglycan cell wall. Their exact function in the survival of T. aquaticus remains unknown but has been theorised to include temporary food and nucleotide storage, or they may play a role in the attachment and organisation of colonies.[5]
T. aquaticus has become famous as a source of thermostable enzymes, particularly the Taq DNA polymerase, as described below.
Studies of this extreme thermophilic bacterium that could be grown in cell culture was initially centered on attempts to understand how enzymes, which are normally inactive at high temperature, can function at high temperature in thermophiles. In 1970, Freeze and Brock published an article describing a thermostable aldolase enzyme from T. aquaticus.[7]
The first polymerase enzyme isolated from T. aquaticus in 1974 was a DNA-dependent RNA polymerase,[8] used in the process of transcription.
Most molecular biologists probably became aware of T. aquaticus in the late 1970s or early 1980s because of the isolation of useful restriction endonucleases from this organism.[9] Use of the term Taq to refer to Thermus aquaticus arose at this time from the convention of giving restriction enzymes short names, such as Sal and Hin, derived from the genus and species of the source organisms.
DNA polymerase was first isolated from T. aquaticus in 1976.[10] The first advantage found for this thermostable (temperature optimum 72°C, does not denature even in 95 °C) DNA polymerase was that it could be isolated in a purer form (free of other enzyme contaminants) than could the DNA polymerase from other sources. Later, Kary Mullis and other investigators at Cetus Corporation discovered this enzyme could be used in the polymerase chain reaction (PCR) process for amplifying short segments of DNA,[11] eliminating the need to add E. coli polymerase enzymes after every cycle of thermal denaturation of the DNA. The enzyme was also cloned, sequenced, modified (to produce the shorter 'Stoffel fragment'), and produced in large quantities for commercial sale.[12] In 1989 Science magazine named Taq polymerase as its first "Molecule of the Year".[13] In 1993, Mullis was awarded the Nobel Prize in Chemistry for his work with PCR.[14]
The high optimum temperature for T. aquaticus allows researchers to study reactions under conditions for which other enzymes lose activity. Other enzymes isolated from this organism include DNA ligase, alkaline phosphatase, NADH oxidase, isocitrate dehydrogenase, amylomaltase, and fructose 1,6-disphosphate-dependent L-lactate dehydrogenase.
The commercial use of enzymes from T. aquaticus has not been without controversy. After Brock's studies, samples of the organism were deposited in the American Type Culture Collection, a public repository. Other scientists, including those at Cetus, obtained it from there. As the commercial potential of Taq polymerase became apparent in the 1990s,[15] the National Park Service labeled its use as the "Great Taq Rip-off".[16] Researchers working in National Parks are now required to sign "benefits sharing" agreements that would send a portion of later profits back to the Park Service.
Thermus aquaticus is a species of bacteria that can tolerate high temperatures, one of several thermophilic bacteria that belong to the Deinococcota phylum. It is the source of the heat-resistant enzyme Taq DNA polymerase, one of the most important enzymes in molecular biology because of its use in the polymerase chain reaction (PCR) DNA amplification technique.